2IQA
 
 | | PFA2 FAB fragment, monoclinic apo form | | Descriptor: | ACETAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IPT
 
 | | PFA1 Fab Fragment | | Descriptor: | ACETAMIDE, IgG2a Fab fragment Heavy Chain, IgG2a Fab fragment Light Chain Kappa | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0Z
 
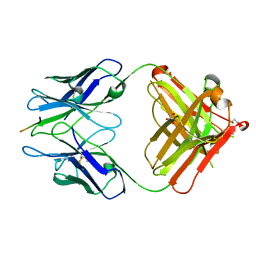 | | PFA1 FAB complexed with GripI peptide fragment | | Descriptor: | GLYCEROL, GripI peptide fragment, IgG2a Fab fragment heavy chain, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0W
 
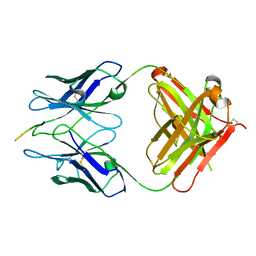 | | PFA2 FAB complexed with Abeta1-8 | | Descriptor: | Amyloid beta peptide fragment, IgG2a Fab fragment heavy chain, Fd portion, ... | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6PF1
 
 | |
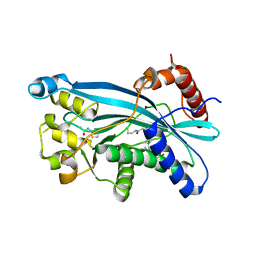
6PGU
 
 | |
3KYU
 
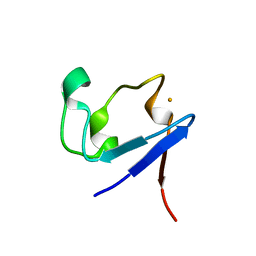 | |
3KYV
 
 | |
3KYX
 
 | |
3KYW
 
 | |
3KYY
 
 | |
4DGQ
 
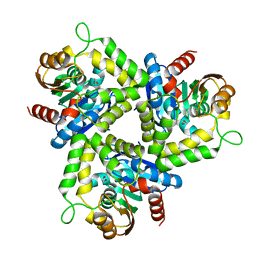 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
2IQ9
 
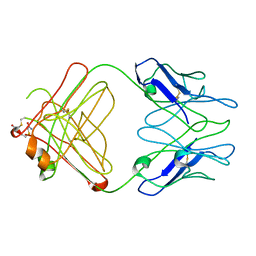 | | PFA2 FAB fragment, triclinic apo form | | Descriptor: | ACETAMIDE, IgG2a Fab fragment PFA2 Kappa light chain, IgG2a Fab fragment PFA2 heavy chain | | Authors: | Gardberg, A.S, Dealwis, C. | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for passive immunotherapy of Alzheimer's disease
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IPU
 
 | |
6V8K
 
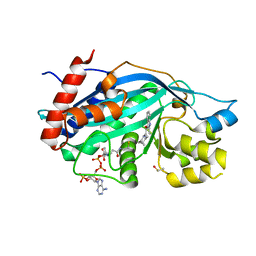 | |
6V8N
 
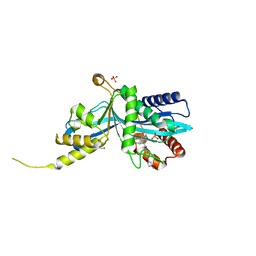 | |
6V8B
 
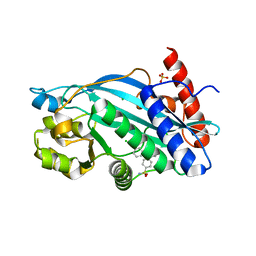 | |
6V90
 
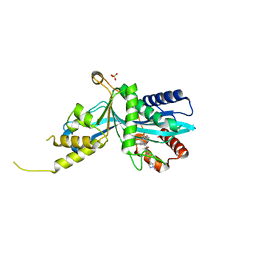 | |
6OMU
 
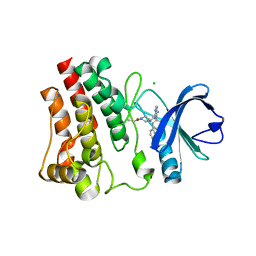 | | Structure of human Bruton's Tyrosine Kinase in complex with Evobrutinib | | Descriptor: | 1-[4-({[6-amino-5-(4-phenoxyphenyl)pyrimidin-4-yl]amino}methyl)piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, Tyrosine-protein kinase BTK | | Authors: | Mochalkin, I, Gardberg, A.S. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Discovery of Evobrutinib: An Oral, Potent, and Highly Selective, Covalent Bruton's Tyrosine Kinase (BTK) Inhibitor for the Treatment of Immunological Diseases.
J.Med.Chem., 62, 2019
|
|
4PJV
 
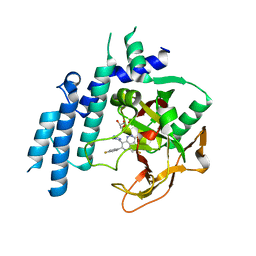 | | Structure of PARP2 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Edwards, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4PJT
 
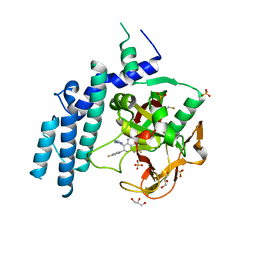 | | Structure of PARP1 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Arakaki, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3EYU
 
 | |
3EYS
 
 | |
4F0L
 
 | |
4FKX
 
 | |
