7QO8
 
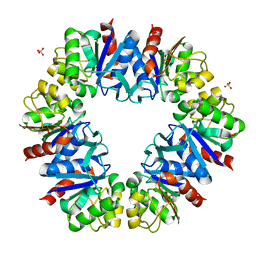 | |
5LDA
 
 | | Structure of deubiquitinating enzyme homolog (Pyrococcus furiosus JAMM1) in complex with ubiquitin-like SAMP2. | | Descriptor: | GLYCEROL, JAMM1, SAMP2, ... | | Authors: | Cao, S, Engilberge, S, Girard, E, Gabel, F, Franzetti, B, Maupin-Furlow, J.A. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
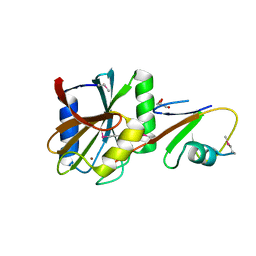
5LD9
 
 | | Structure of deubiquitinating enzyme homolog, Pyrococcus furiosus JAMM1. | | Descriptor: | CHLORIDE ION, JAMM1, ZINC ION | | Authors: | Maupin-Furlow, J.A, Franzetti, B, Cao, S, Girard, E, Gabel, F, Engilberge, S. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-17 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
2XEB
 
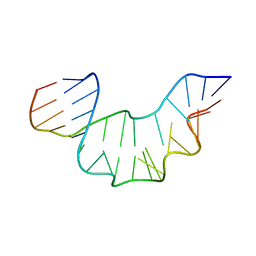 | | NMR STRUCTURE OF THE PROTEIN-UNBOUND SPLICEOSOMAL U4 SNRNA 5' STEM LOOP | | Descriptor: | 5'-R(P*GP*AP*UP*CP*GP*UP*AP*GP*CP*CP*AP*AP*UP*GP*AP* GP*GP*UP*U)-3', 5'-R(P*GP*CP*CP*GP*AP*GP*GP*CP*GP*CP*GP*AP*UP*C)-3' | | Authors: | Falb, M, Amata, I, Gabel, F, Simon, B, Carlomagno, T. | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the K-turn U4 RNA: a combined NMR and SANS study.
Nucleic Acids Res., 38, 2010
|
|
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
7ANU
 
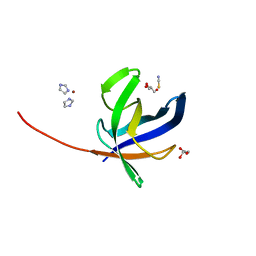 | | Crystal structure of Pyrococcus abyssi Pbp11 | | Descriptor: | GLYCEROL, IMIDAZOLE, NICKEL (II) ION, ... | | Authors: | Hogrel, G, Girard, E, Franzetti, B. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization of a small tRNA-binding protein that interacts with the archaeal proteasome complex.
Mol.Microbiol., 118, 2022
|
|
2WZN
 
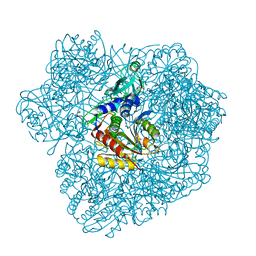 | | 3d structure of TET3 from Pyrococcus horikoshii | | Descriptor: | 354AA LONG HYPOTHETICAL OPERON PROTEIN FRV, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rosenbaum, E, Dura, M.A, Vellieux, F.M, Franzetti, B. | | Deposit date: | 2009-12-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural and Biochemical Characterizations of a Novel Tet Peptidase Complex from Pyrococcus Horikoshii Reveal an Integrated Peptide Degradation System in Hyperthermophilic Archaea.
Mol.Microbiol., 72, 2009
|
|
6O22
 
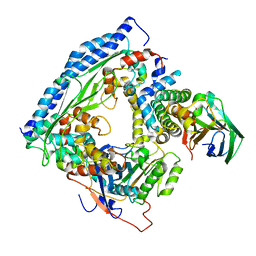 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
6DCL
 
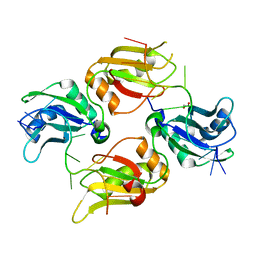 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
5EQT
 
 | |
3MS6
 
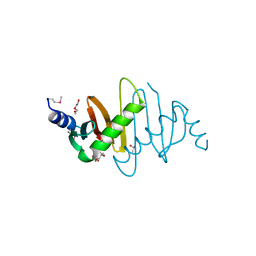 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
3MSH
 
 | |
5O3J
 
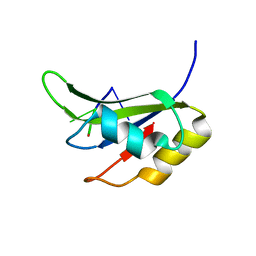 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | Descriptor: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | Authors: | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O2V
 
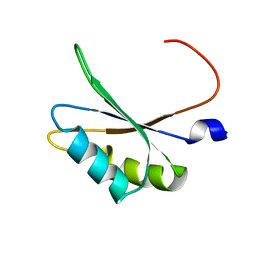 | | NMR structure of TIA-1 RRM1 domain | | Descriptor: | Nucleolysin TIA-1 isoform p40 | | Authors: | Jagtap, P.K.A. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3ZFJ
 
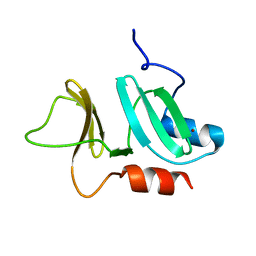 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
