3R0L
 
 | | Crystal structure of crotoxin | | Descriptor: | ACETATE ION, CHLORIDE ION, Crotoxin chain A, ... | | Authors: | Saul, F.A, Faure, G. | | Deposit date: | 2011-03-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Crotoxin Reveals Key Residues Involved in the Stability and Toxicity of This Potent Heterodimeric Beta-Neurotoxin
J.Mol.Biol., 412, 2011
|
|
3G8H
 
 | |
8EEX
 
 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
8EEY
 
 | | Cas7-11 in complex with DR-mismatched target RNA, Csx29 and Csx30 | | Descriptor: | Cas7-11, Csx29, Csx30, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
6TMY
 
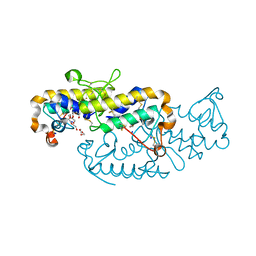 | | Crystal structure of isoform CBd of the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus | | Descriptor: | CHLORIDE ION, Phospholipase A2 crotoxin basic subunit CBc, SODIUM ION, ... | | Authors: | Nemecz, D, Ostrowski, M, Saul, F.A, Faure, G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Isoform CBd of the Basic Phospholipase A 2 Subunit of Crotoxin: Description of the Structural Framework of CB for Interaction with Protein Targets.
Molecules, 25, 2020
|
|
2J4W
 
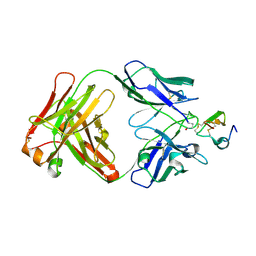 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
2J5L
 
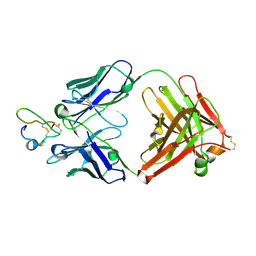 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
3G8G
 
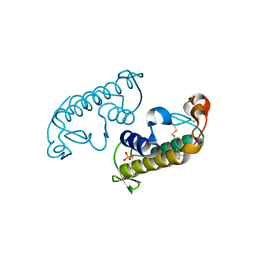 | |
3Q4F
 
 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
9CF0
 
 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*GP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*TP*AP*TP*CP*CP*CP*GP*GP*GP*T)-3'), DNA/RNA (54-MER), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CET
 
 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CER
 
 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, Guillardia theta Fanzor1 chimera, RNA (142-MER) | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEZ
 
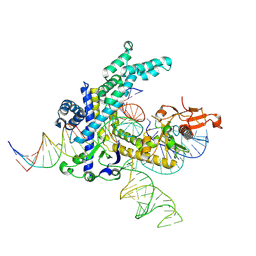 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEX
 
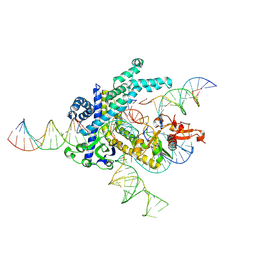 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEY
 
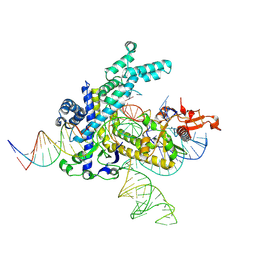 | |
9CF3
 
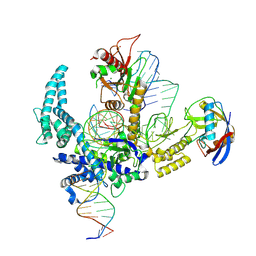 | | Parasitella parasitica Fanzor (PpFz) State 4 | | Descriptor: | DNA (31-MER), DNA (5'-D(P*AP*CP*CP*CP*GP*GP*GP*TP*AP*TP*A)-3'), DNA/RNA (56-MER), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEV
 
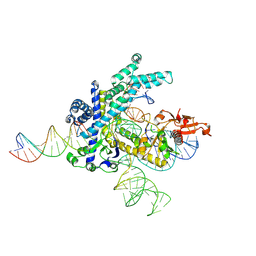 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEW
 
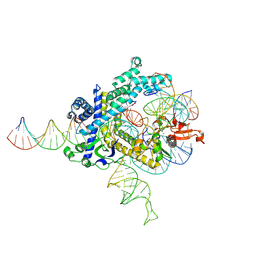 | | Spizellomyces punctatus Fanzor (SpuFz) State 3 | | Descriptor: | DNA (29-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | Descriptor: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CF1
 
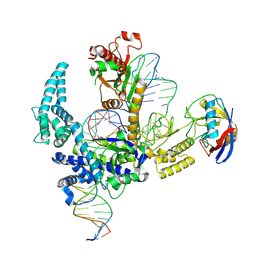 | | Parasitella parasitica Fanzor (PpFz) State 2 | | Descriptor: | DNA (5'-D(P*AP*CP*CP*CP*GP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(P*GP*CP*TP*GP*GP*AP*TP*GP*TP*TP*TP*AP*TP*CP*CP*CP*GP*GP*GP*T)-3'), DNA/RNA (51-MER), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CES
 
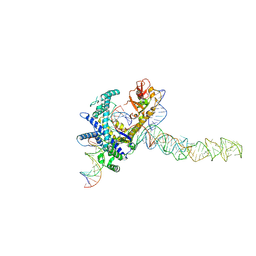 | | Guillardia theta Fanzor (GtFz) State 2 | | Descriptor: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
9CF2
 
 | | Parasitella parasitica Fanzor (PpFz) State 3 | | Descriptor: | DNA (26-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 2024
|
|
8GKH
 
 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2023-03-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
8UYO
 
 | |
8DMB
 
 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
