256L
 
 | | BACTERIOPHAGE T4 LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Faber, H.R, Matthews, B.W. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A mutant T4 lysozyme displays five different crystal conformations.
Nature, 348, 1990
|
|
1VFE
 
 | | HUMAN LACTOFERRIN, N-TERMINAL LOBE MUTANT WITH ARG 121 REPLACED BY SER (R121S) | | Descriptor: | CARBONATE ION, FE (III) ION, HUMAN LACTOFERRIN | | Authors: | Faber, H.R, Day, C.L, Baker, E.N. | | Deposit date: | 1996-10-01 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutation of arginine 121 in lactoferrin destabilizes iron binding by disruption of anion binding: crystal structures of R121S and R121E mutants.
Biochemistry, 35, 1996
|
|
1VFD
 
 | | HUMAN LACTOFERRIN, N-TERMINAL LOBE MUTANT WITH ARG 121 REPLACED BY GLU (R121E) | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Faber, H.R, Day, C.L, Baker, E.N. | | Deposit date: | 1996-10-01 | | Release date: | 1997-04-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of arginine 121 in lactoferrin destabilizes iron binding by disruption of anion binding: crystal structures of R121S and R121E mutants.
Biochemistry, 35, 1996
|
|
1HXN
 
 | |
1DSN
 
 | | D60S N-TERMINAL LOBE HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Faber, H.R, Norris, G.E, Baker, E.N. | | Deposit date: | 1995-12-13 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered domain closure and iron binding in transferrins: the crystal structure of the Asp60Ser mutant of the amino-terminal half-molecule of human lactoferrin.
J.Mol.Biol., 256, 1996
|
|
150L
 
 | |
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-10-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1SGH
 
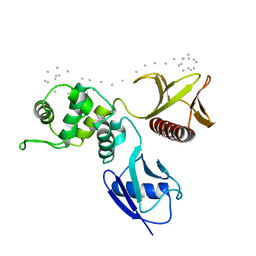 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | Descriptor: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | Authors: | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
3DK8
 
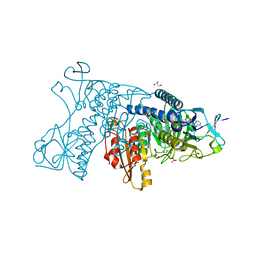 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
3DK9
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, SULFATE ION | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
3DJJ
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Glutathione reductase, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
3DK4
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, Glutathione reductase, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
3DJG
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
1KNI
 
 | | Stabilizing Disulfide Bridge Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Jacobson, R.H, Matsumura, M, Faber, H.R, Matthews, B.W. | | Deposit date: | 2001-12-18 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a stabilizing disulfide bridge mutant that closes the active-site cleft of T4 lysozyme.
Protein Sci., 1, 1992
|
|
1CGO
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1CGN
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1GLA
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, GLYCEROL KINASE | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
1GLB
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, ... | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
3LVB
 
 | | Crystal structure of the Ferredoxin:NADP+ reductase from maize root at 1.7 angstroms - Test Set Withheld | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues
Biochemistry, 40, 2001
|
|
1JB9
 
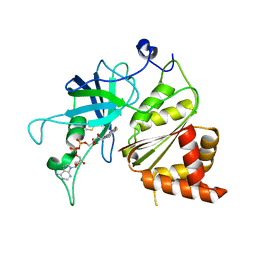 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
