2G7L
 
 | | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor | | Descriptor: | TetR-family transcriptional regulator | | Authors: | Ezersky, A, Lunin, V.V, Skarina, T, Wierzbicka, M, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor
To be Published
|
|
1YSQ
 
 | | The crystal structure of transcriptional regulator YaiJ | | Descriptor: | HTH-type transcriptional regulator yiaJ, PHOSPHATE ION | | Authors: | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
1YSP
 
 | | Crystal structure of the C-terminal domain of E. coli transcriptional regulator KdgR. | | Descriptor: | SULFATE ION, Transcriptional regulator kdgR | | Authors: | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
2O9A
 
 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2O99
 
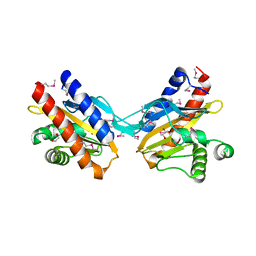 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
1TF1
 
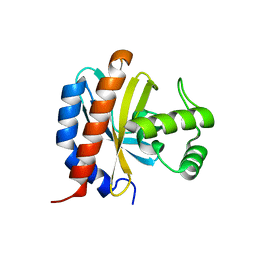 | | Crystal Structure of the E. coli Glyoxylate Regulatory Protein Ligand Binding Domain | | Descriptor: | Negative regulator of allantoin and glyoxylate utilization operons | | Authors: | Walker, J.R, Skarina, T, Kudrytska, M, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical study of effector molecule recognition by the E.coli glyoxylate and allantoin utilization regulatory protein AllR.
J.Mol.Biol., 358, 2006
|
|
