8I4T
 
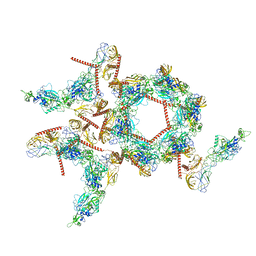 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
3M48
 
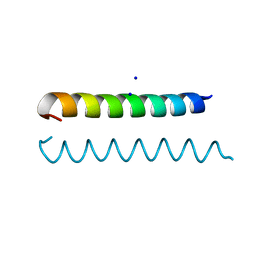 | | GCN4 Leucine Zipper Peptide Mutant | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Du, S, Kettering, R.D, Alvarado, J.J, Tortajada, A, Yeh, J.I. | | Deposit date: | 2010-03-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | GCN4 Leucine Zipper Peptide Mutant
To be Published
|
|
7CHC
 
 | |
8SPL
 
 | | Proteinase K Multiconformer Model at 343K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOV
 
 | | Proteinase K Multiconformer Model at 353K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOG
 
 | | Proteinase K Multiconformer Model at 313K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOU
 
 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SQV
 
 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
7CH5
 
 | |
7CH4
 
 | |
8ILQ
 
 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
5Y92
 
 | | Crystal structure of ANXUR2 extracellular domain from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-like protein kinase ANXUR2, ... | | Authors: | Du, S, Xiao, J.Y. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR2
Protein Sci., 27, 2018
|
|
5Y96
 
 | | Crystal structure of ANXUR1 extracellular domain from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-like protein kinase ANXUR1 | | Authors: | Du, S, Xiao, J.Y. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR2
Protein Sci., 27, 2018
|
|
7WCH
 
 | |
7UY0
 
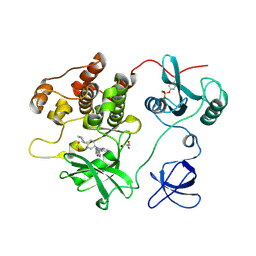 | | Crystal structure of human Fgr tyrosine kinase in complex with A-419259 | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
7UY3
 
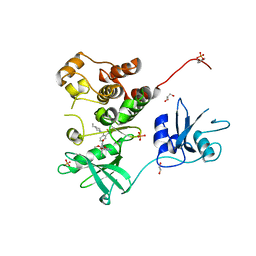 | | Crystal structure of human Fgr tyrosine kinase in complex with TL02-59 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6,7-dimethoxyquinazolin-4-yl)oxy]-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, ... | | Authors: | Du, S, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | ATP-site inhibitors induce unique conformations of the acute myeloid leukemia-associated Src-family kinase, Fgr.
Structure, 30, 2022
|
|
7WCU
 
 | |
7WCK
 
 | |
7WCP
 
 | |
7WR9
 
 | |
7V20
 
 | | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Du, S, Xiao, J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | CryoEM structure of del68-76/del679-688 prefusion-stabilized spike
To Be Published
|
|
7V22
 
 | |
7V23
 
 | |
7V24
 
 | |
7DXI
 
 | |
