6RGL
 
 | |
6RAP
 
 | |
6RAO
 
 | |
6RC8
 
 | |
6RBK
 
 | |
6RBN
 
 | |
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H5S
 
 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4UFT
 
 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
9E0Q
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei D-hydrazino-Lysine analog at 2.3 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
5AHV
 
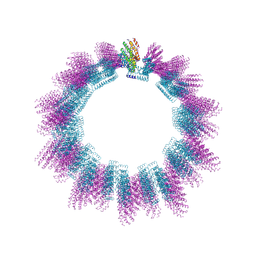 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YN6
 
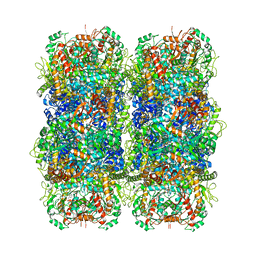 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PK6
 
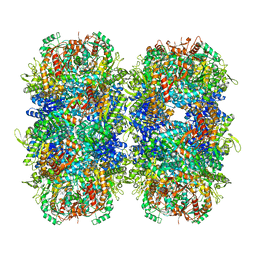 | | Providencia stuartii Arginine decarboxylase (Adc), stack structure | | Descriptor: | Biodegradative arginine decarboxylase | | Authors: | Jessop, M, Desfosses, A, Bacia-Verloop, M, Gutsche, I. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation.
Commun Biol, 5, 2022
|
|
7P9B
 
 | | Providencia stuartii Arginine decarboxylase (Adc), decamer structure | | Descriptor: | Biodegradative arginine decarboxylase | | Authors: | Jessop, M, Desfosses, A, Bacia-Verloop, M, Gutsche, I. | | Deposit date: | 2021-07-26 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural and biochemical characterisation of the Providencia stuartii arginine decarboxylase shows distinct polymerisation and regulation.
Commun Biol, 5, 2022
|
|
8OP2
 
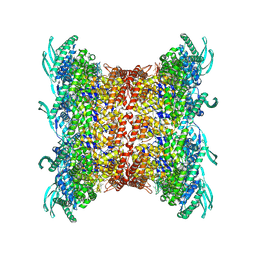 | |
8OP1
 
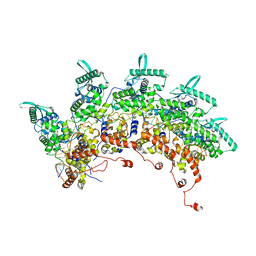 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
8OOU
 
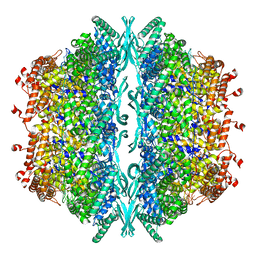 | |
8ALY
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament (G1032W mutant) | | Descriptor: | Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Mariotti, L, Inian, O, Desfosses, A, Beuron, F, Morris, E.P, Guettler, S. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of tankyrase activation by polymerization.
Nature, 612, 2022
|
|
7ZCH
 
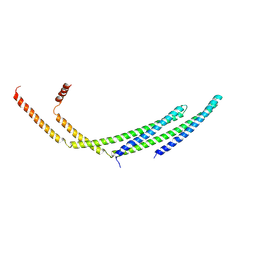 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZCG
 
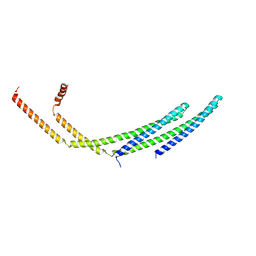 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9E0M
 
 | | CryoEM structure of holoenzyme of inducible Lysine decarboxylase from Hafnia alvei holoenzyme at 2.19 Angstrom resolution | | Descriptor: | Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
9E0O
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei L-hydrazino-Lysine analog at 2.04 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
8S5C
 
 | | Cryo-EM structure of Arf1-decorated membrane tubules | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosylation factor 1, MAGNESIUM ION | | Authors: | Haupt, C, Semchonok, D.A, Stubbs, M.T, Bacia, K, Desfosses, A, Kastritis, P.L, Hamdi, F. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Arf1-decorated membrane tubules
To Be Published
|
|
8S5D
 
 | | Cryo-EM structure of Arf1-decorated membrane tubules | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosylation factor 1, MAGNESIUM ION | | Authors: | Haupt, C, Semchonok, D.A, Stubbs, M.T, Bacia, K, Desfosses, A, Kastritis, P.L, Hamdi, F. | | Deposit date: | 2024-02-23 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structure of Arf1-decorated membrane tubules
To Be Published
|
|
