3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1C
 
 | | Crystal structure of de novo designed MID1-zinc | | Descriptor: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1B
 
 | | Crystal structure of de novo designed MID1-apo2 | | Descriptor: | Computational design, MID1-apo2, GLYCEROL | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
 | | Crystal structure of de novo designed MID1-cobalt | | Descriptor: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
5KAY
 
 | | Structure of Spelter bound to Zn2+ | | Descriptor: | SODIUM ION, Spelter, ZINC ION | | Authors: | Guffy, S.L, Der, B.S, Kuhlman, B. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-03 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the minimal determinants of zinc binding with computational protein design.
Protein Eng.Des.Sel., 29, 2016
|
|
5K7J
 
 | |
2LSE
 
 | | Solution NMR Structure of De Novo Designed Four Helix Bundle Protein, Northeast Structural Genomics Consortium (NESG) Target OR188 | | Descriptor: | Four Helix Bundle Protein | | Authors: | Sathyamoorthy, B, Pulavarti, S, Murphy, G, Mills, J.L, Eletski, A, Der, B.S, Machius, M.C, Kuhlman, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Denovo Design Four Helix Bundle Protein, Northeast Structural Genomics Consortium Target OR188
To be Published
|
|
6P79
 
 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
5OD1
 
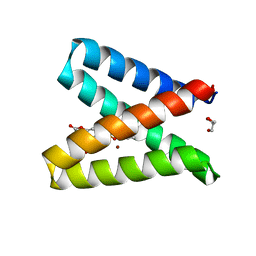 | | Structure of the engineered metalloesterase MID1sc10 complexed with a phosphonate transition state analogue | | Descriptor: | GLYCEROL, MID1sc10, ZINC ION, ... | | Authors: | Mittl, P.R.E, Studer, S, Hansen, D.A, Hilvert, D. | | Deposit date: | 2017-07-04 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
5OD9
 
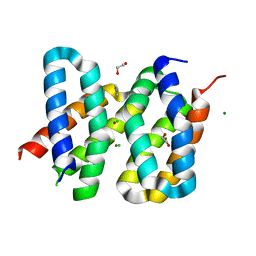 | | Structure of the engineered metalloesterase MID1sc9 | | Descriptor: | (2~{S})-2-phenylpropanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Studer, S, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2017-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
1MKB
 
 | |
1MKA
 
 | |
