6DKQ
 
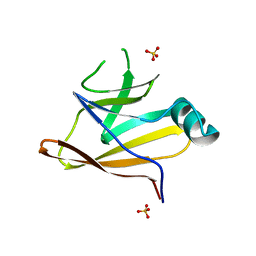 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | Descriptor: | Heme-binding protein Shr, SULFATE ION | | Authors: | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
5K2H
 
 | |
5K2F
 
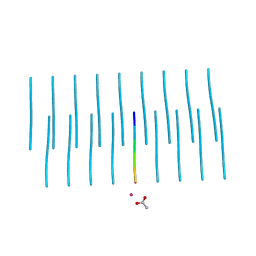 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZK7
 
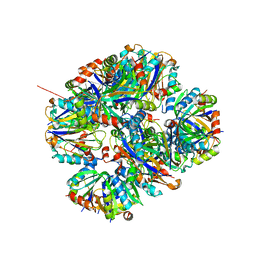 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
5IM5
 
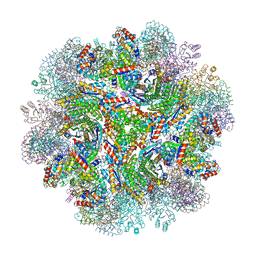 | | Crystal structure of designed two-component self-assembling icosahedral cage I53-40 | | Descriptor: | Designed Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase, Designed Riboflavin synthase | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.699 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
5IM6
 
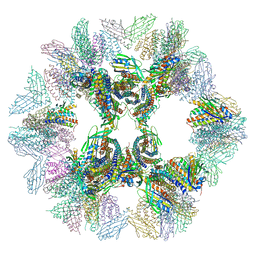 | | Crystal structure of designed two-component self-assembling icosahedral cage I32-28 | | Descriptor: | Designed self-assembling icosahedral cage I32-28 dimeric subunit, Designed self-assembling icosahedral cage I32-28 trimeric subunit | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.588 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
5IM4
 
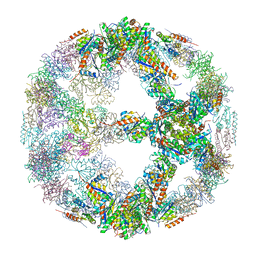 | | Crystal structure of designed two-component self-assembling icosahedral cage I52-32 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, Phosphotransferase system, mannose/fructose-specific component IIA | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
