6FUY
 
 | | Crystal structure of human full-length vinculin-T12-A974K (residues 1-1066) | | Descriptor: | CALCIUM ION, Vinculin | | Authors: | Chorev, D.S, Volberg, T, Livne, A, Eisenstein, M, Martins, B, Kam, Z, Jockusch, B.M, Medalia, O, Sharon, M, Geiger, B. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational states during vinculin unlocking differentially regulate focal adhesion properties.
Sci Rep, 8, 2018
|
|
5FQ8
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-decanoyloxypropyl decanoate, BT_2262 (UNCHARACTERISED LIPOPROTEIN), ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ7
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2261, BT_2262, BT_2263, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ6
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | 3-decanoyloxypropyl decanoate, BT_2261, CALCIUM ION, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ4
 
 | | Crystal structure of the lipoprotein BT2263 from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, PUTATIVE LIPOPROTEIN | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ3
 
 | | Crystal structure of the lipoprotein BT2262 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2262 | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5LX8
 
 | | Crystal structure of BT1762 | | Descriptor: | SODIUM ION, SULFATE ION, SusD homolog | | Authors: | Basle, A. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
6I3Y
 
 | | Crystal structure of the human mitochondrial PRELID1K58V-TRIAP1 complex with PS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, PRELI domain-containing protein 1, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I4Y
 
 | | X-ray structure of the human mitochondrial PRELID3b-TRIAP1 complex | | Descriptor: | Maltose transport system, substrate-binding protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3B, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I3V
 
 | | x-ray structure of the human mitochondrial PRELID1 in complex with TRIAP1 | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, PRELI domain-containing protein 1, ... | | Authors: | Berry, J.L, Miliara, X, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6SRS
 
 | | Structure of the Fanconi anaemia core subcomplex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (central region, proposed FANCB-FAAP100), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
6SRI
 
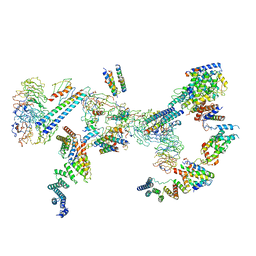 | | Structure of the Fanconi anaemia core complex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (base region, proposed FANCC-FANC-E-FANCF), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
6MHY
 
 | |
6MHQ
 
 | |
5O65
 
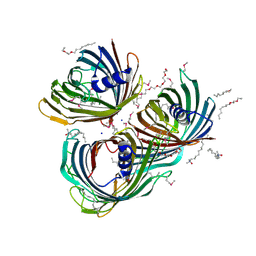 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O67
 
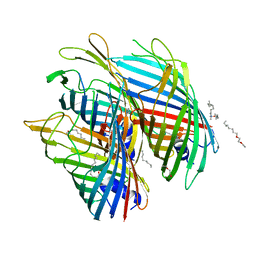 | | Crystal structure of the FapF polypeptide transporter - F103A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O68
 
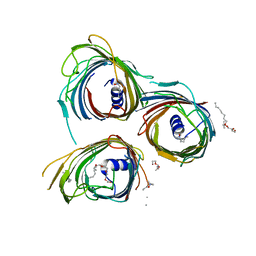 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF - R157A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5T4Y
 
 | | Crystal structure of BT1762-1763 | | Descriptor: | MAGNESIUM ION, SusC homolog, SusD homolog | | Authors: | van den Berg, B. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5T3R
 
 | | Crystal structure of BT1762-1763 | | Descriptor: | MAGNESIUM ION, SODIUM ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2016-08-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
