4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1E
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
3RVB
 
 | | The structure of HCV NS3 helicase (Heli-80) bound with inhibitor ITMN-3479 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA helicase, ... | | Authors: | Mather, O, Cheng, R, Schonfield, D, Barker, J. | | Deposit date: | 2011-05-06 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of HCV NS3 helicase with inhibitor ITMN-3479
To be Published
|
|
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
9HC0
 
 | | Dark structure of the human metabotropic glutamate receptor 5 transmembrane domain bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin, OLEIC ACID | | Authors: | Kondo, Y, Hatton, C, Cheng, R, Trabuco, M, Glover, H, Bertrand, Q, Stierli, F, Seidel, H.P, Mason, T, Sarma, S, Tellkamp, F, Kepa, M, Dworkowski, F, Mehrabi, P, Hennig, M, Standfuss, J. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Apo-state structure of the metabotropic glutamate receptor 5 transmembrane domain obtained using a photoswitchable ligand.
Protein Sci., 34, 2025
|
|
9HC3
 
 | | Apo-state structure of the human metabotropic glutamate receptor 5 transmembrane domain freeze-trapped after light activation of photoswitchable ligand alloswitch-1 | | Descriptor: | Metabotropic glutamate receptor 5,Endolysin, OLEIC ACID | | Authors: | Kondo, Y, Hatton, C, Cheng, R, Trabuco, M, Glover, H, Bertrand, Q, Stierli, F, Seidel, H.P, Mason, T, Sarma, S, Tellkamp, F, Kepa, M, Dworkowski, F, Mehrabi, P, Hennig, M, Standfuss, J. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Apo-state structure of the metabotropic glutamate receptor 5 transmembrane domain obtained using a photoswitchable ligand.
Protein Sci., 34, 2025
|
|
7LMA
 
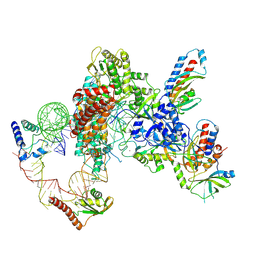 | | Tetrahymena telomerase T3D2 structure at 3.3 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMB
 
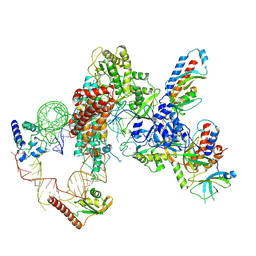 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
5YM0
 
 | | The crystal structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, SULFATE ION | | Authors: | Zang, X, Huang, W.X, Cheng, R, Wu, L, Zhou, J.H, Tang, Y, Yan, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The crystal structure of DHAD
To Be Published
|
|
3FHR
 
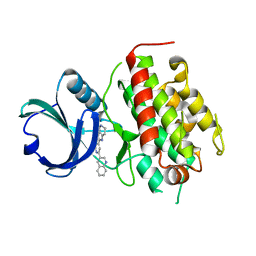 | | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3 (MK3)-inhibitor complex | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Cheng, R.K.Y, Barker, J, Palan, S, Felicetti, B, Whittaker, M, Hesterkamp, T. | | Deposit date: | 2008-12-10 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of human Mapkap kinase 3 in complex with a high affinity ligand
Protein Sci., 19, 2010
|
|
6KTW
 
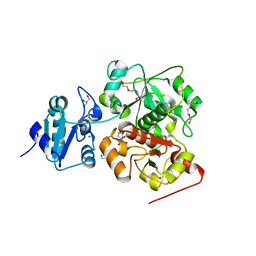 | | structure of EanB with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KU2
 
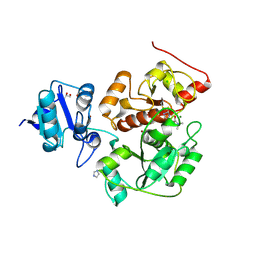 | | The structure of EanB/Y353A complex with ergothioneine covalent linked with persulfide Cys412 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTZ
 
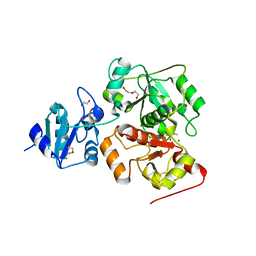 | | The complex structure of EanB/C412S with hercynine | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTV
 
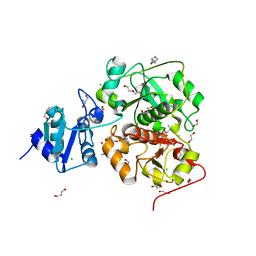 | | The structure of EanB complex with hercynine and persulfided Cys412 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTX
 
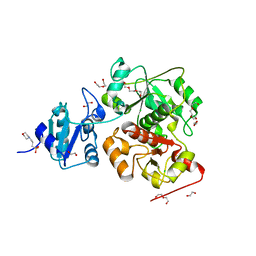 | | The wildtype structure of EanB | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KU1
 
 | | The structure of EanB/Y353A complex with ergothioneine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
