1XOB
 
 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1XOA
 
 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1ZNF
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
6WXH
 
 | | Colicin E1 fragment in nanodisc-embedded TolC | | Descriptor: | Colicin-E1, Outer membrane protein TolC | | Authors: | Kaelber, J.T, Budiardjo, S.J, Firlar, E, Case, D.A, Ikujuni, A.P, Slusky, J.S.G. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Colicin E1 opens its hinge to plug TolC.
Elife, 11, 2022
|
|
9CBX
 
 | | Tetrahymena ribozyme with automatically identified water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 642, 2025
|
|
9CBW
 
 | | Tetrahymena ribozyme with consensus water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 642, 2025
|
|
9CBU
 
 | | Tetrahymena ribozyme with consensus water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 642, 2025
|
|
9CBY
 
 | | Tetrahymena ribozyme with automatically identified water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 642, 2025
|
|
8U3M
 
 | | The FARFAR-MD-NMR ensemble of an HIV-1 TAR excited state | | Descriptor: | The excited state of HIV-1 transactivation response element (31-MER) | | Authors: | Geng, A, Ganser, L, Roy, R, Shi, H, Pratihar, S, Case, D.A, Al-Hashimi, H.M. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA excited conformational state at atomic resolution.
Nat Commun, 14, 2023
|
|
1TF3
 
 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1AX3
 
 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1HV2
 
 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
1CQO
 
 | |
2HSK
 
 | | NMR Structure of 13mer Duplex DNA containing an abasic site (Y) in 5'-CCAAAGYACCGGG-3' (10 structures, alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HPX
 
 | | 13mer Duplex DNA containing a 4'-oxidized abasic site, averaged structure | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(X4A)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HOU
 
 | | Structure ensembles of duplex DNA containing a 4'-oxidized abasic site. | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(X4A)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-16 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
1MYF
 
 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
2HSR
 
 | | 13mer duplex DNA containing an abasic site with beta anomer | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HSL
 
 | | NMR structure of 13mer duplex DNA containing an abasic site, averaged structure (alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HSS
 
 | | 13mer duplex DNA containg an abasic site with beta anomer, averaged structure | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
1DSM
 
 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
1DSA
 
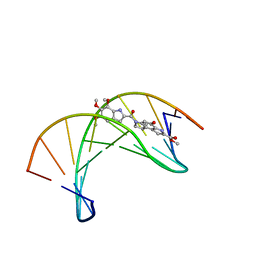 | | (+)-DUOCARMYCIN SA COVALENTLY LINKED TO DUPLEX DNA, NMR, 20 STRUCTURES | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Eis, P.S, Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA.
J.Mol.Biol., 272, 1997
|
|
1DGQ
 
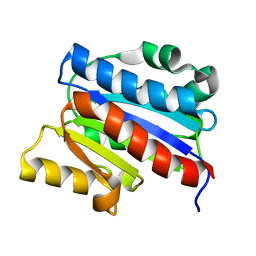 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
1PQQ
 
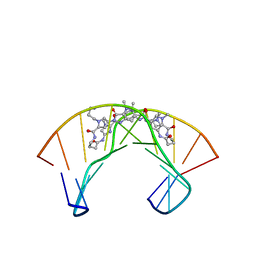 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
