1C3Q
 
 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
4THI
 
 | | THIAMINASE I FROM BACILLUS THIAMINOLYTICUS WITH COVALENTLY BOUND 4-AMINO-2,5-DIMETHYLPYRIMIDINE | | Descriptor: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, PROTEIN (THIAMINASE I), SULFATE ION | | Authors: | Campobasso, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thiaminase-I from Bacillus thiaminolyticus at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
2THI
 
 | |
1ESQ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1ESJ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) | | Descriptor: | HYDROXYETHYLTHIAZOLE KINASE, SULFATE ION | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
3THI
 
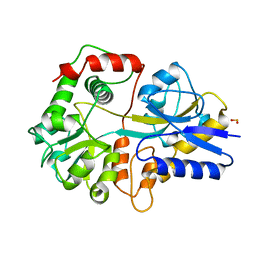 | |
1EKK
 
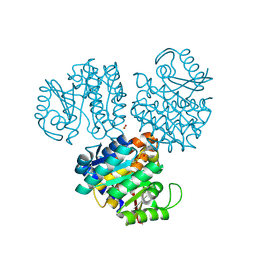 | | CRYSTAL STRUCTURE OF HYDROXYETHYLTHIAZOLE KINASE IN THE R3 FORM WITH HYDROXYETHYLTHIAZOLE | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, HYDROXYETHYLTHIAZOLE KINASE, SULFUR DIOXIDE | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-09 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1EKQ
 
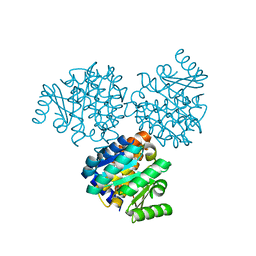 | |
1TXT
 
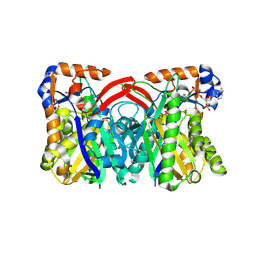 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TVZ
 
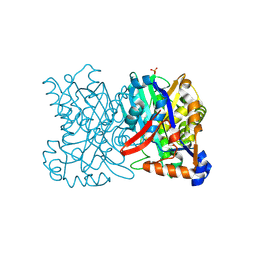 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
7LCU
 
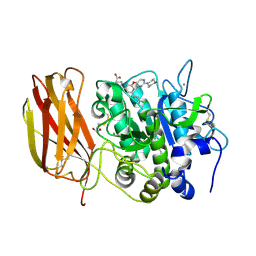 | | X-ray structure of Furin bound to BOS-318, a small molecule inhibitor | | Descriptor: | (1-{[2-(3,5-dichlorophenyl)-6-{[2-(4-methylpiperazin-1-yl)pyrimidin-5-yl]oxy}pyridin-4-yl]methyl}piperidin-4-yl)acetic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Campobasso, N, Reid, R. | | Deposit date: | 2021-01-11 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A highly selective, cell-permeable furin inhibitor BOS-318 rescues key features of cystic fibrosis airway disease.
Cell Chem Biol, 29, 2022
|
|
5TX5
 
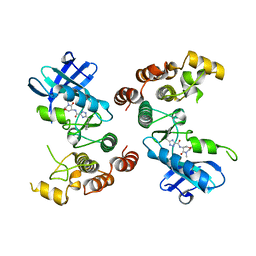 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
6N4T
 
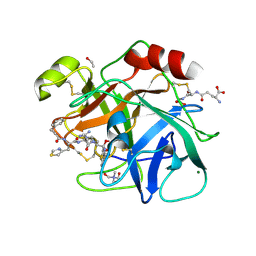 | |
6NPE
 
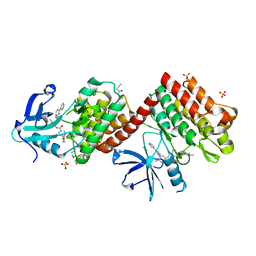 | | C-abl Kinase domain with the activator(cmpd6), 2-cyano-N-(4-(3,4-dichlorophenyl)thiazol-2-yl)acetamide | | Descriptor: | 2-cyano-~{N}-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]ethanamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, NONAETHYLENE GLYCOL, ... | | Authors: | campobasso, N. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and Optimization of Novel Small c-Abl Kinase Activators Using Fragment and HTS Methodologies.
J. Med. Chem., 62, 2019
|
|
6NPV
 
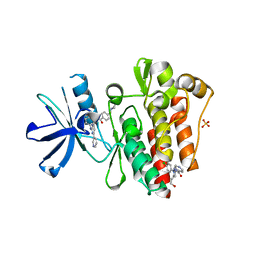 | |
6NPU
 
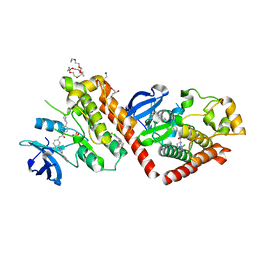 | |
5HFU
 
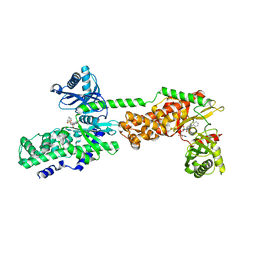 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HEX
 
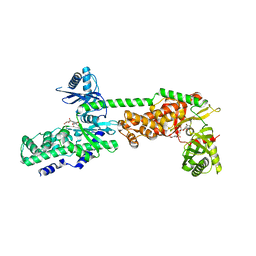 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HG1
 
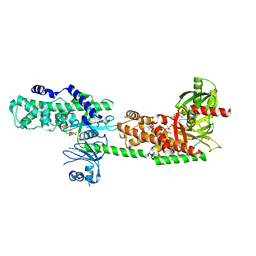 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HX6
 
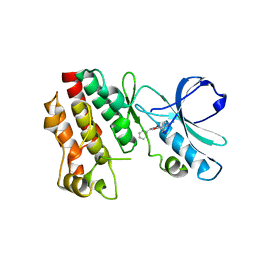 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6OW2
 
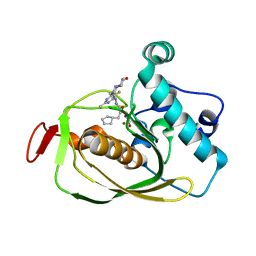 | | X-ray Structure of Polypeptide Deformylase | | Descriptor: | (2R)-2-(cyclopentylmethyl)-N'-{5-fluoro-6-[(9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]-2-methylpyrimidin-4-yl}-3-[hydroxy(hydroxymethyl)amino]propanehydrazide, NICKEL (II) ION, Peptide deformylase | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OW7
 
 | | X-ray Structure of Polypeptide Deformylase with a Piperazic Acid | | Descriptor: | (3S)-2-{(2R)-2-(cyclopentylmethyl)-3-[formyl(hydroxy)amino]propanoyl}-N-(pyridin-2-yl)hexahydropyridazine-3-carboxamide, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3SW8
 
 | |
3SVJ
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine amide | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-methyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-12 | | Release date: | 2011-07-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
3STR
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine hydroxamic acid | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-[2-(hydroxyamino)-2-oxoethyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
