1BRS
 
 | |
1BSE
 
 | |
1BSA
 
 | |
1BSD
 
 | |
1BSB
 
 | |
1BRN
 
 | |
1BSC
 
 | |
1KID
 
 | |
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
2DG4
 
 | | FK506-binding protein mutant WF59 complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2DG3
 
 | | Wildtype FK506-binding protein complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
1JON
 
 | |
1BNI
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1CIQ
 
 | | COMPLEX OF TWO FRAGMENTS OF CI2, RESIDUES 1-40 AND 41-64 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Buckle, A.M, Fersht, A.R. | | Deposit date: | 1995-10-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Towards the complete structural characterization of a protein folding pathway: the structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism
Structure Fold.Des., 1, 1996
|
|
3G01
 
 | |
3FZZ
 
 | | Structure of GrC | | Descriptor: | Granzyme C, SULFATE ION | | Authors: | Buckle, A.M, Kaiserman, D, Whisstock, J.C. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of granzyme C reveals an unusual mechanism of protease autoinhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H7O
 
 | |
3H7T
 
 | |
2GPZ
 
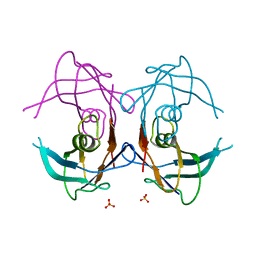 | |
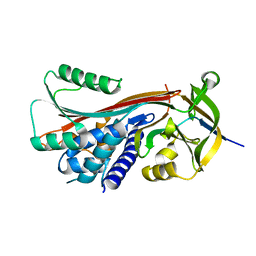
2H4P
 
 | |
2OKK
 
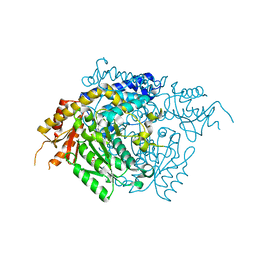 | | The X-ray crystal structure of the 65kDa isoform of Glutamic Acid Decarboxylase (GAD65) | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, Glutamate decarboxylase 2 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OKJ
 
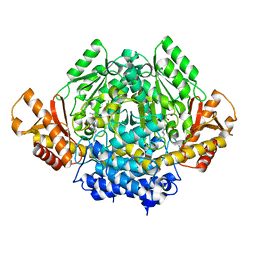 | | The X-ray crystal structure of the 67kDa isoform of Glutamic Acid Decarboxylase (GAD67) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase 1 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1B27
 
 | |
1B3S
 
 | |
1B2S
 
 | |
