4V7E
 
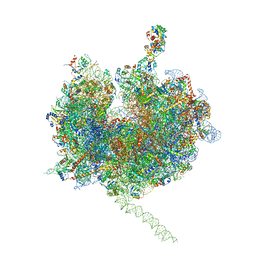 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | Authors: | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JPQ
 
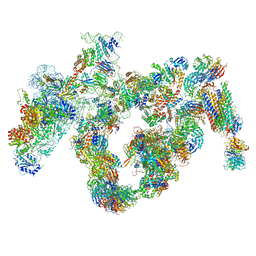 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6TNU
 
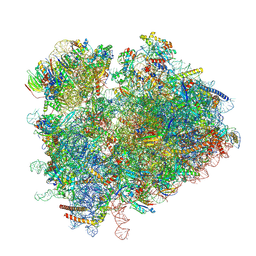 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6Q8Y
 
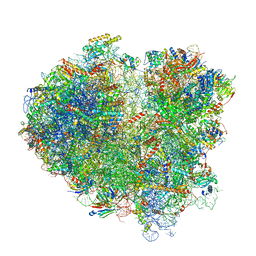 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5JCS
 
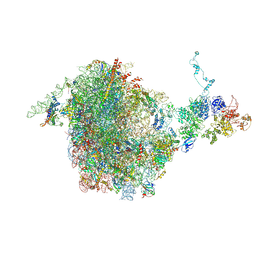 | | CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Barrio-Garcia, C, Thoms, M, Flemming, D, Kater, L, Berninghausen, O, Bassler, J, Beckmann, R, Hurt, E. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling
Nat.Struct.Mol.Biol., 23, 2016
|
|
8BIP
 
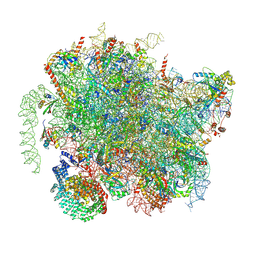 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BJQ
 
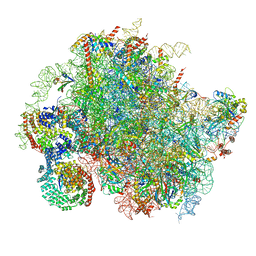 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6T83
 
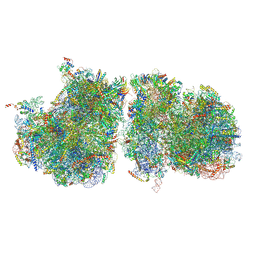 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7T
 
 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
4V6M
 
 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4V6X
 
 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
4V6W
 
 | | Structure of the D. melanogaster 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
4UY8
 
 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Bischoff, L, Berninghausen, O, Beckmann, R. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
5A8L
 
 | | Human eRF1 and the hCMV nascent peptide in the translation termination complex | | Descriptor: | 28S RIBOSOMAL RNA, 60S RIBOSOMAL PROTEIN L12, 60S RIBOSOMAL PROTEIN L17, ... | | Authors: | Matheisl, S, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Human Translation Termination Complex.
Nucleic Acids Res., 43, 2015
|
|
5ABB
 
 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6EML
 
 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
