1P4W
 
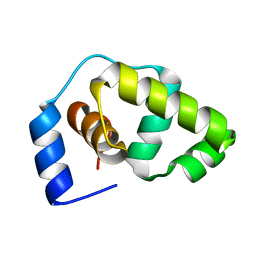 | | Solution structure of the DNA-binding domain of the Erwinia amylovora RcsB protein | | Descriptor: | rcsB | | Authors: | Pristovsek, P, Sengupta, K, Loehr, F, Schaefer, B, Wehland von Trebra, M, Rueterjans, H, Bernhard, F. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box.
J.Biol.Chem., 278, 2003
|
|
1SR2
 
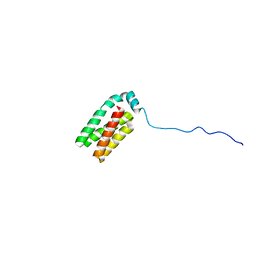 | |
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
2RON
 
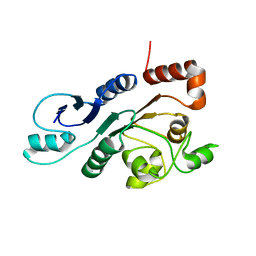 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2016-10-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
8POK
 
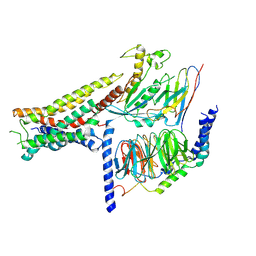 | | Cryo-EM structure of cell-free synthesized human histamine H2 receptor coupled to heterotrimeric Gs protein in lipid environment | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HISTAMINE, ... | | Authors: | Schnelle, K, Koeck, Z, Persechino, M, Umbach, S, Schihada, H, Januliene, D, Parey, K, Pockes, S, Kolb, P, Doetsch, V, Moeller, A, Hilger, D, Bernhard, F. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Nat Commun, 15, 2024
|
|
2AYY
 
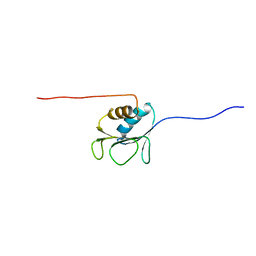 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
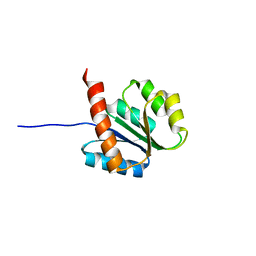 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYX
 
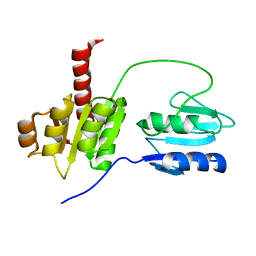 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2KX7
 
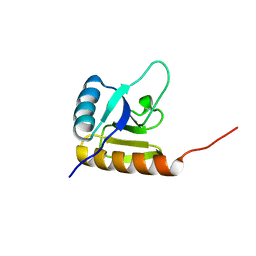 | | Solution structure of the E.coli RcsD-ABL domain (residues 688-795) | | Descriptor: | Sensor-like histidine kinase yojN | | Authors: | Rogov, V.V, Schmoee, K, Rogova, N.Y, Loehr, F, Bernhard, F, Doetsch, V. | | Deposit date: | 2010-04-27 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Rcs Phosphotransfer: The Newly Identified RcsD-ABL Domain Enhances Interaction with the Response Regulator RcsB.
Structure, 19, 2011
|
|
2L6X
 
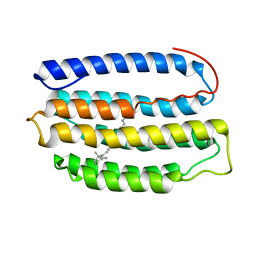 | | Solution NMR Structure of Proteorhodopsin. | | Descriptor: | Green-light absorbing proteorhodopsin, RETINAL | | Authors: | Reckel, S, Gottstein, D, Stehle, J, Loehr, F, Takeda, M, Silvers, R, Kainosho, M, Glaubitz, C, Bernhard, F, Schwalbe, H, Guntert, P, Doetsch, V, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-09 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of proteorhodopsin.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2GDX
 
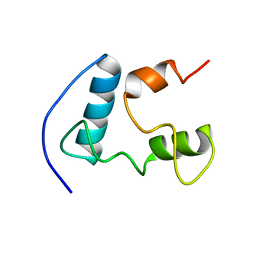 | | Solution structure of the B. brevis TycC3-PCP in H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2GDW
 
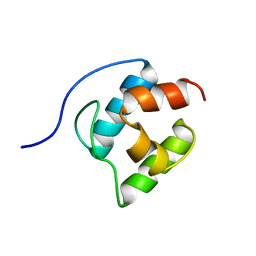 | | Solution structure of the B. brevis TycC3-PCP in A/H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2GDY
 
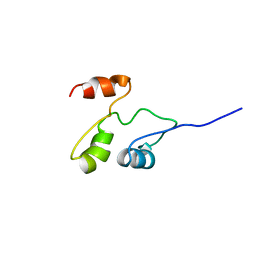 | | Solution structure of the B. brevis TycC3-PCP in A-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2K2Q
 
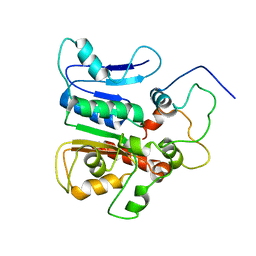 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
5J9P
 
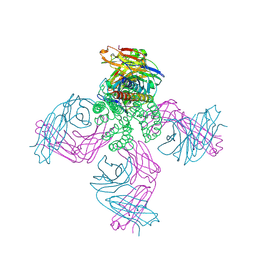 | | KcsA in vitro | | Descriptor: | Fab, POTASSIUM ION, pH-gated potassium channel KcsA | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2016-04-10 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining in Vitro Folding with Cell Free Protein Synthesis for Membrane Protein Expression.
Biochemistry, 55, 2016
|
|
3SJB
 
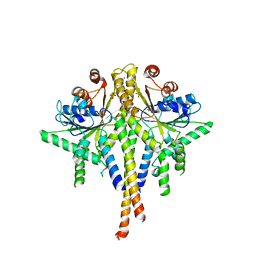 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJC
 
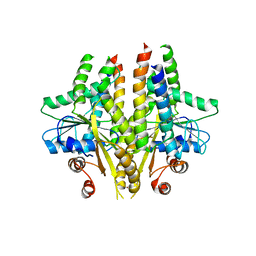 | |
3SJA
 
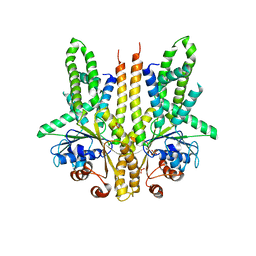 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJD
 
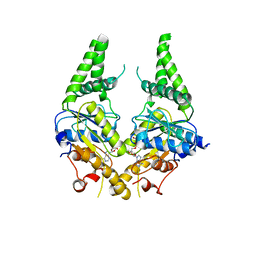 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
5BR2
 
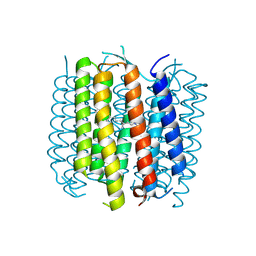 | | Structure of bacteriorhodopsin crystallized from ND-MSP1 | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nikolaev, M, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-05-29 | | Release date: | 2016-09-14 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integral Membrane Proteins Can Be Crystallized Directly from Nanodiscs
Cryst.Growth Des., 17, 2017
|
|
5BR5
 
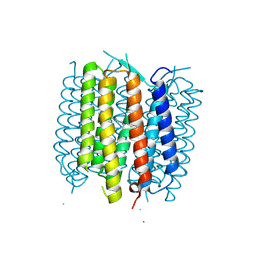 | | Structure of bacteriorhodopsin crystallized from ND-MSP1E3D1 | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nikolaev, M, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-05-29 | | Release date: | 2016-09-14 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Integral Membrane Proteins Can Be Crystallized Directly from Nanodiscs
Cryst.Growth Des., 17, 2017
|
|
4BPD
 
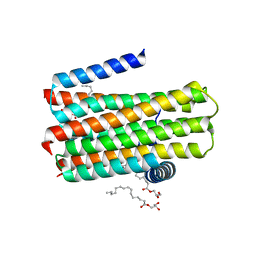 | | Structure determination of an integral membrane kinase | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, DIACYLGLYCEROL KINASE, ZINC ION | | Authors: | Li, D, Boland, C, Caffrey, M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cell-Free Expression and in Meso Crystallisation of an Integral Membrane Kinase for Structure Determination.
Cell.Mol.Life Sci., 71, 2014
|
|
4D2E
 
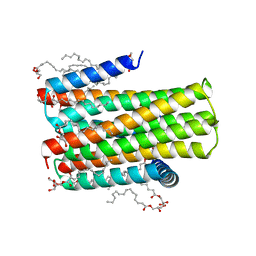 | | Crystal structure of an integral membrane kinase - v2.3 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, CITRATE ANION, ... | | Authors: | Li, D, Boland, C, Caffrey, M. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cell-Free Expression and in Meso Crystallisation of an Integral Membrane Kinase for Structure Determination.
Cell.Mol.Life Sci., 71, 2014
|
|
