1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Authors: | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
8S5N
 
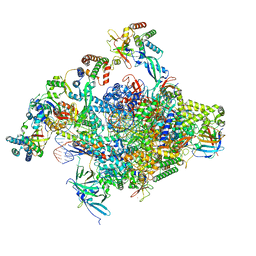 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S55
 
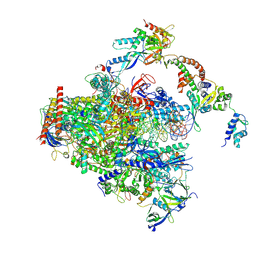 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
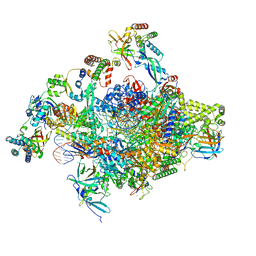 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S52
 
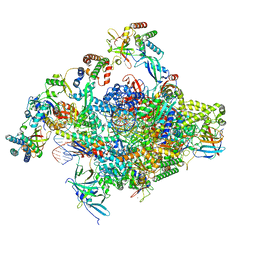 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S54
 
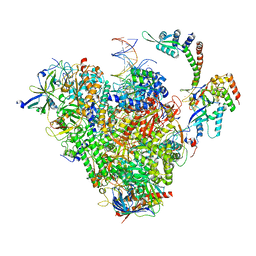 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
9GD3
 
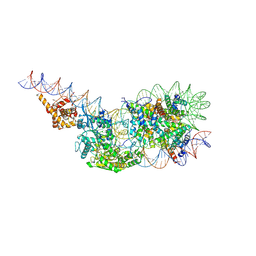 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD1
 
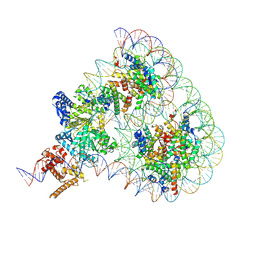 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD2
 
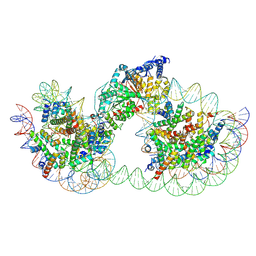 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
9GD0
 
 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of Transcription-Induced Hexasome-Nucleosome Complexes by Chd1 and FACT.
Mol.Cell, 2024
|
|
3TTJ
 
 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1BRD
 
 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
8CCZ
 
 | |
8CCW
 
 | |
5NBL
 
 | | Crystal structure of the Arp4-N-actin(APO-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5NBN
 
 | | Crystal structure of the Arp4-N-actin-Arp8-Ino80HSA module of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7MJ9
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived mutant IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, Insulin-like growth factor-binding protein-like 1 altered peptide, MHC class I antigen | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ6
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ7
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJA
 
 | | HLA-A*24:02 bound to Neuroblastoma derived PHOX2B peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ8
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
5NBM
 
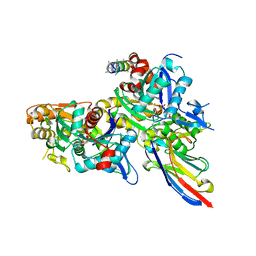 | | Crystal structure of the Arp4-N-actin(ATP-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1AT9
 
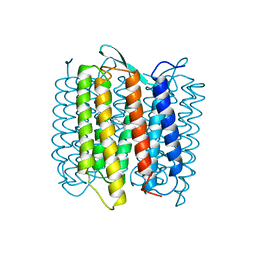 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
1TEY
 
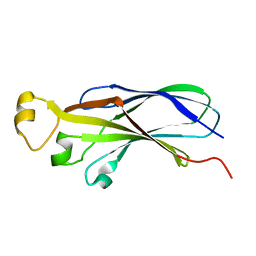 | | NMR structure of human histone chaperone, ASF1A | | Descriptor: | ASF1 anti-silencing function 1 homolog A | | Authors: | Mousson, F, Lautrette, A, Thuret, J.Y, Agez, M, Amigues, B, Courbeyrette, R, Neumann, J.M, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2004-05-26 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of Asf1 with histone H3 and its functional implications.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2BRD
 
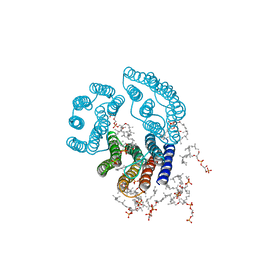 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN PURPLE MEMBRANE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Henderson, R, Grigorieff, N. | | Deposit date: | 1995-12-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Electron-crystallographic refinement of the structure of bacteriorhodopsin.
J.Mol.Biol., 259, 1996
|
|
