2X0V
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2X0W
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 5,6-dimethoxy- 2-methylbenzothiazole | | Descriptor: | 5,6-DIMETHOXY-2-METHYL-1,3-BENZOTHIAZOLE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Kaar, J.L, Basse, N, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2X0U
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
3NZW
 
 | | Crystal structure of the yeast 20S proteasome in complex with 2b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3NZJ
 
 | | Crystal structure of yeast 20S proteasome in complex with ligand 2a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-16 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3NZX
 
 | | Crystal structure of the yeast 20S proteasome in complex with ligand 2c | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C97
 
 | | Crystal structure of FcRn at pH3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | Authors: | Fox III, D, Fairman, J.W. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
2L9N
 
 | |
6AX6
 
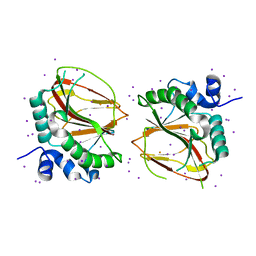 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
6AX7
 
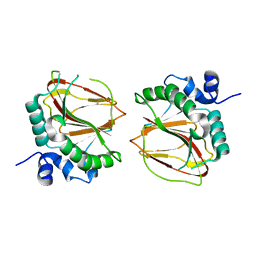 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
