4B1F
 
 | | Design of Inhibitors of Helicobacter pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty | | Descriptor: | 5-methyl-3-(1-methyl-1H-imidazol-5-yl)-7-(2-methylpropyl)-2-(naphthalen-1-ylmethyl)-2H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Basarab, G.S, Hill, P, Eyermann, C.J, Gowravaram, M, Kack, H, Osimoni, E. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Inhibitors of Helicobacter Pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles Onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4LP0
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
6STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 3 | | Descriptor: | 2,2-DICHLORO-1-METHANESULFINYL-3-METHYL-CYCLOPROPANECARBOXYLIC ACID [1-(4-BROMO-PHENYL)-ETHYL]-AMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4STD
 
 | | HIGH RESOLUTION STRUCTURES OF SCYTALONE DEHYDRATASE-INHIBITOR COMPLEXES CRYSTALLIZED AT PHYSIOLOGICAL PH | | Descriptor: | N-[1-(4-BROMOPHENYL)ETHYL]-5-FLUORO SALICYLAMIDE, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
5STD
 
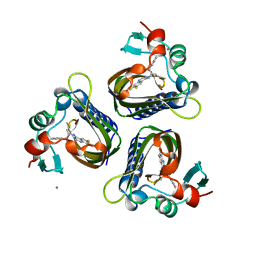 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 2 | | Descriptor: | (6,7-DIFLUORO-QUINAZOLIN-4-YL)-(1-METHYL-2,2-DIPHENYL-ETHYL)-AMINE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4WF1
 
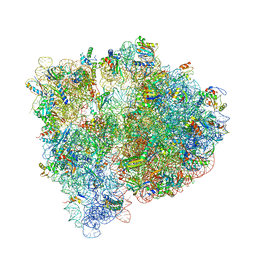 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3STD
 
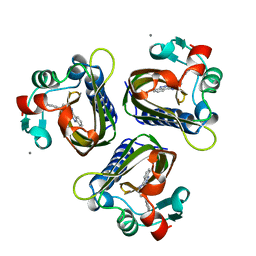 | | SCYTALONE DEHYDRATASE AND CYANOCINNOLINE INHIBITOR | | Descriptor: | 4-(3,3-diphenylpropylamino)cinnoline-3-carbonitrile, CALCIUM ION, PROTEIN (SCYTALONE DEHYDRATASE) | | Authors: | Chen, J.M, Xu, S.L, Wawrzak, Z, Basarab, G.S, Jordan, D.B. | | Deposit date: | 1998-10-16 | | Release date: | 1999-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based design of potent inhibitors of scytalone dehydratase: displacement of a water molecule from the active site.
Biochemistry, 37, 1998
|
|
7STD
 
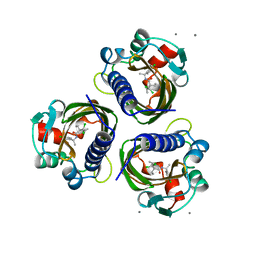 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
1DOH
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND 4-NITRO-INDEN-1-ONE | | Descriptor: | 4-NITRO-INDEN-1-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D.I, Basarab, G.S, Gatenby, A.A, Jordan, D.B. | | Deposit date: | 1999-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
1G0N
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND 4,5,6,7-TETRACHLORO-PHTHALIDE | | Descriptor: | 4,5,6,7-TETRACHLORO-PHTHALIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D, Basarab, G.S, Gatenby, A.A, Valent, B, Jordan, D.B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
1G0O
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND PYROQUILON | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROQUILON, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D, Basarab, G.S, Gatenby, A.A, Valent, B, Jordan, D.B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
4LPB
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 1-ethyl-3-{5'-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridin-6-yl}urea, Topoisomerase IV subunit B | | Authors: | Boriack-Sjodin, A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
1IDP
 
 | |
1STD
 
 | |
7Q26
 
 | | Crystal structure of Angiotensin-1 converting enzyme N-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
7Q29
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD013 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2021-10-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
7Q24
 
 | |
7Q25
 
 | |
7Q28
 
 | |
7Q27
 
 | |
1ASK
 
 | | NUCLEAR TRANSPORT FACTOR 2 (NTF2) H66A MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Mccoy, A.J, Stewart, M.J. | | Deposit date: | 1997-08-11 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran.
J.Mol.Biol., 272, 1997
|
|
1AR0
 
 | | NUCLEAR TRANSPORT FACTOR 2 (NTF2) E42K MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Mccoy, A.J, Stewart, M.J. | | Deposit date: | 1997-08-08 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran.
J.Mol.Biol., 272, 1997
|
|
1OUN
 
 | |
