4R2Y
 
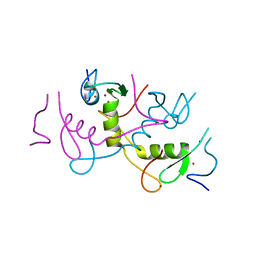 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
3A0E
 
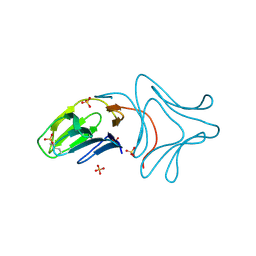 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
3A0C
 
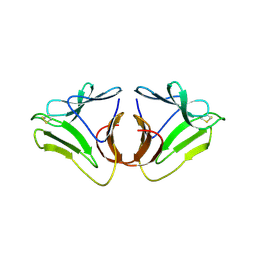 | |
3A0D
 
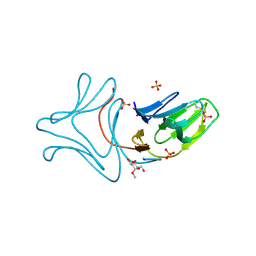 | |
7SSB
 
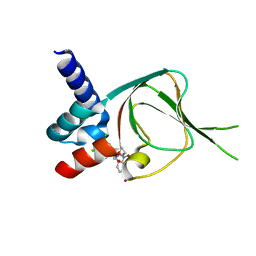 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
5XJ7
 
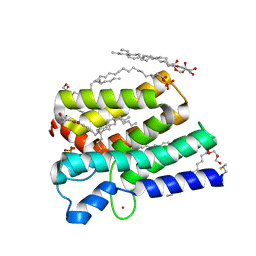 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the acyl phosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Tang, Y, Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ9
 
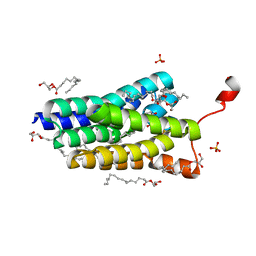 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the orthophosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ5
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the monoacylglycerol form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLYCINE, Glycerol-3-phosphate acyltransferase, ... | | Authors: | Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
4KR0
 
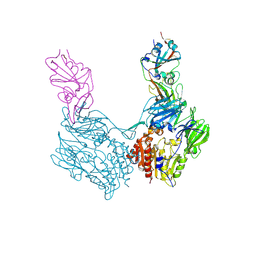 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
2MAH
 
 | | Solution structure of Smoothened | | Descriptor: | Protein smoothened | | Authors: | Rana, R, Lee, H, Zheng, J.J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling.
Nat Commun, 4, 2013
|
|
2MT5
 
 | | Isolated Ring domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissman, F, Royappa, G, Schulman, B, Jarvis, M, Vanderlinden, R, Frye, J.J, Qiao, R, Petzold, G, Peters, J, Stark, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
5XJ8
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ6
 
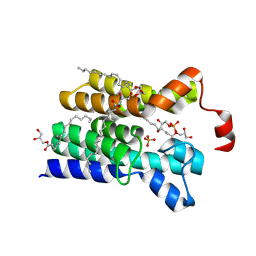 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the glycerol 3-phosphate form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
7FIF
 
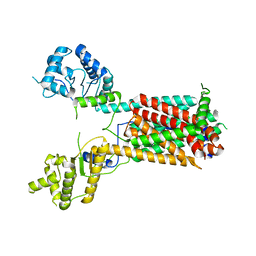 | | Cryo-EM structure of the hedgehog release protein Disp from water bear (Hypsibius dujardini) | | Descriptor: | Protein dispatched-like protein 1 | | Authors: | Luo, Y, Wan, G, Wang, Q, Zhao, Y, Cong, Y, Li, D. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture of Dispatched, a Transmembrane Protein Responsible for Hedgehog Release.
Front Mol Biosci, 8, 2021
|
|
4E5E
 
 | | Crystal structure of avian influenza virus PAn Apo | | Descriptor: | MANGANESE (II) ION, Polymerase protein PA, SULFATE ION | | Authors: | DuBois, R.M, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5G
 
 | | Crystal structure of avian influenza virus PAn bound to compound 2 | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5F
 
 | | Crystal structure of avian influenza virus PAn bound to compound 1 | | Descriptor: | 2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5L
 
 | | Crystal structure of avian influenza virus PAn bound to compound 6 | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5J
 
 | | Crystal structure of avian influenza virus PAn bound to compound 5 | | Descriptor: | 2-[3-(acetylamino)phenyl]-5-hydroxy-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5I
 
 | | Crystal structure of avian influenza virus PAn bound to compound 4 | | Descriptor: | 5-hydroxy-2-(1-methyl-1H-imidazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5H
 
 | | Crystal structure of avian influenza virus PAn bound to compound 3 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
