3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
2R04
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R07
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(6-CHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R06
 
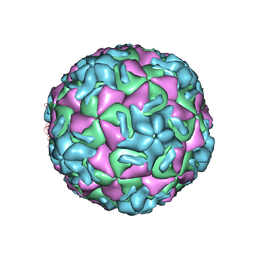 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS5
 
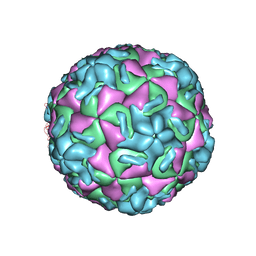 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RM2
 
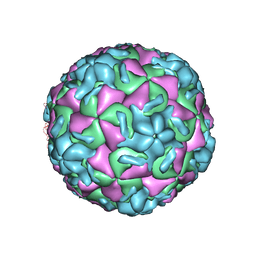 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(6-CHLORO-4-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS3
 
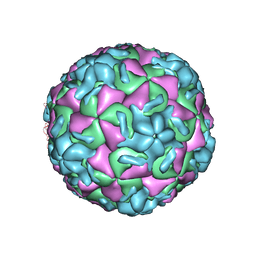 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-ETHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RR1
 
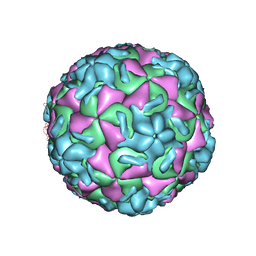 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2RS1
 
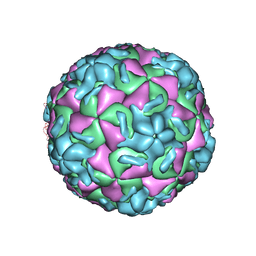 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
1R08
 
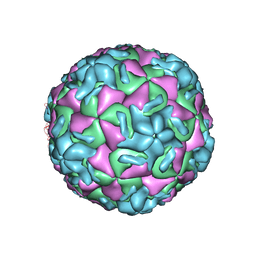 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-(HYDROXYETHYL OXYMETHYLENEOXYMETHYL) ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
1RMU
 
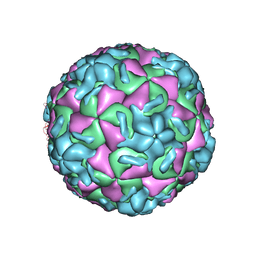 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
4E79
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.66A resolution (P4322 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4E6T
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E6U
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E75
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.85A resolution (P21 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
9INS
 
 | |
3TVX
 
 | | The structure of PDE4A with pentoxifylline at 2.84A resolution | | Descriptor: | 3,7-DIMETHYL-1-(5-OXOHEXYL)-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Badger, J, Sridhar, V. | | Deposit date: | 2011-09-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Fragment-Based Screening for Inhibitors of PDE4A Using Enthalpy Arrays and X-ray Crystallography.
J Biomol Screen, 17, 2012
|
|
4RQR
 
 | | Crystal Structure of Human Glutaredoxin with MESNA | | Descriptor: | 1-THIOETHANESULFONIC ACID, Glutaredoxin-1 | | Authors: | Badger, J, Sridhar, V, Logan, C, Hausheer, F.H, Nienaber, V.L. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Cysteine Specific Targeting of the Functionally Distinct Peroxiredoxin and Glutaredoxin Proteins by the Investigational Disulfide BNP7787.
Molecules, 20, 2015
|
|
4RQX
 
 | | Crystal structure of human peroxiredoxin 4(THIOREDOXIN PEROXIDASE) with MESNA | | Descriptor: | 1-THIOETHANESULFONIC ACID, Peroxiredoxin-4 | | Authors: | Badger, J, Sridhar, V, Logan, C, Hausheer, F.H, Nienaber, V.L. | | Deposit date: | 2014-11-05 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Cysteine Specific Targeting of the Functionally Distinct Peroxiredoxin and Glutaredoxin Proteins by the Investigational Disulfide BNP7787.
Molecules, 20, 2015
|
|
4TT7
 
 | | Crystal structure of human ALK with a covalent modification | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ALK tyrosine kinase receptor | | Authors: | Badger, J, Sridhar, V, Chie-Leon, B, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-06-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel covalent modification of human anaplastic lymphoma kinase (ALK) and potentiation of crizotinib-mediated inhibition of ALK activity by BNP7787.
Onco Targets Ther, 8, 2015
|
|
1XBC
 
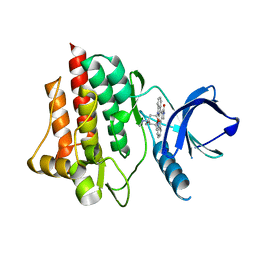 | | Crystal structure of the syk tyrosine kinase domain with Staurosporin | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase SYK | | Authors: | Badger, J, Atwell, S, Adams, J.M, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase
J.Biol.Chem., 279, 2004
|
|
1DPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 11), INSULIN B CHAIN (PH 11), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1CPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN (PH 10), SODIUM ION | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
