6I2N
 
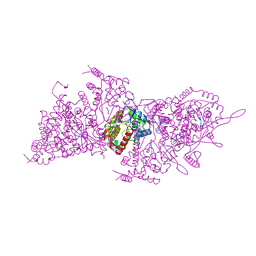 | | Helical RNA-bound Hantaan virus nucleocapsid | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*U)-3') | | Authors: | Arragain, B, Reguera, J, Desfosses, A, Gutsche, I, Schoehn, G, Malet, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Elife, 8, 2019
|
|
6Z8K
 
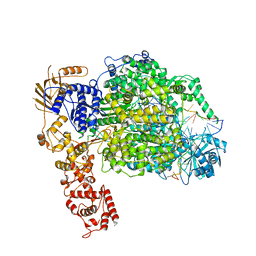 | | La Crosse virus polymerase at elongation mimicking stage | | Descriptor: | La Crosse virus 3' vRNA (1-16), La Crosse virus 5' vRNA (9-16), La Crosse virus 5' vRNA 1-10, ... | | Authors: | Arragain, B, Effantin, G, Schoehn, G, Cusack, S, Malet, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
6Z6G
 
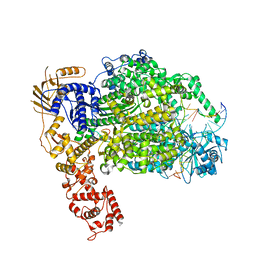 | | Cryo-EM structure of La Crosse virus polymerase at pre-initiation stage | | Descriptor: | 3'vRNA 1-16, 5'vRNA 1-10, 5'vRNA 9-16, ... | | Authors: | Arragain, B, Effantin, G, Gerlach, P, Reguera, J, Schoehn, G, Cusack, S, Malet, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
7ORJ
 
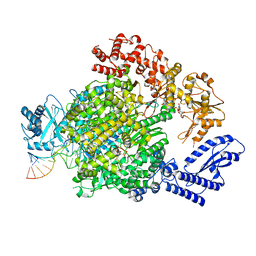 | | La Crosse virus polymerase at transcription capped RNA cleavage stage | | Descriptor: | 5' capped RNA, ADENOSINE-5'-TRIPHOSPHATE, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORI
 
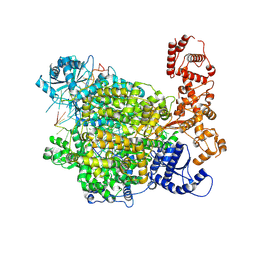 | | La Crosse virus polymerase at replication late-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORN
 
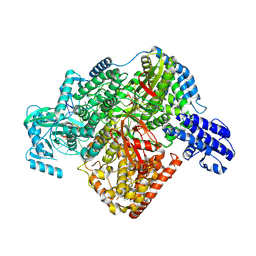 | | La Crosse virus polymerase at replication initiation stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, La Crosse virus polymerase, MAGNESIUM ION, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORL
 
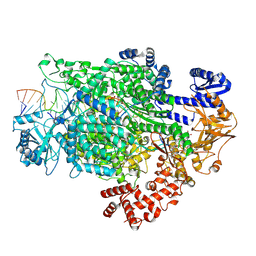 | | La Crosse virus polymerase at transcription initiation stage | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORM
 
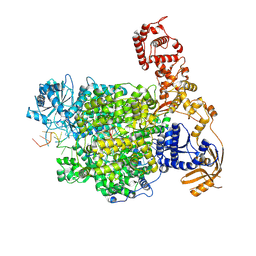 | | La Crosse virus polymerase at transcription early-elongation stage | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORK
 
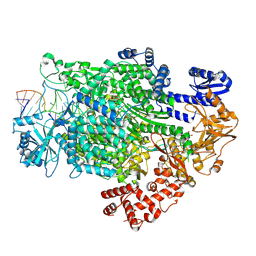 | | La Crosse virus polymerase in transcription mode with cleaved capped RNA entering the polymerase active site | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-D(*(GTG))-R(P*A)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORO
 
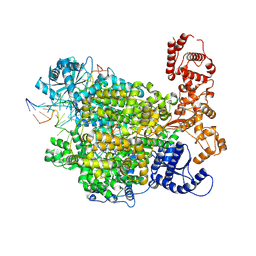 | | La Crosse virus polymerase at replication early-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
9HBS
 
 | | TiLV-NP tetramer (pseudo-C2) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBR
 
 | |
9HBW
 
 | | TiLV-NP tetramer (pseudo-C4) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBZ
 
 | | TiLV-NP hexamer (pseudo-C6) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBV
 
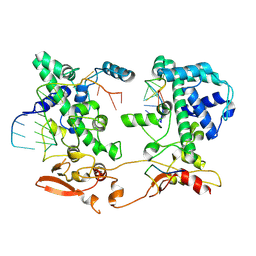 | |
9HBU
 
 | |
9HBY
 
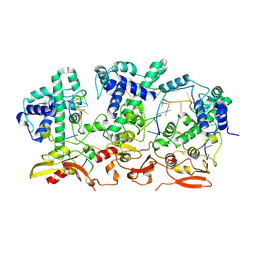 | |
9HBX
 
 | |
9HBT
 
 | | TiLV-NP pentamer (pseudo-C5) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
8RMP
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core1) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RN3
 
 | |
8RN5
 
 | |
8RN8
 
 | | Influenza B polymerase pseudo-symmetrical apo-dimer (FluPol(E)|FluPol(S)) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RMQ
 
 | | Influenza polymerase A/H7N9-4M (ENDO(R) | Core2) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structures of influenza A and B replication complexes give insight into avian to human host adaptation and reveal a role of ANP32 as an electrostatic chaperone for the apo-polymerase.
Nat Commun, 15, 2024
|
|
8RMR
 
 | |
