5K79
 
 | | Structure and anti-HIV activity of CYT-CVNH, a new cyanovirin-n homolog | | Descriptor: | 1,2-ETHANEDIOL, Cyanovirin-N domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Matei, E, Basu, R, Furey, W, Shi, J, Calnan, C, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Glycan Binding of a New Cyanovirin-N Homolog.
J.Biol.Chem., 291, 2016
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
3LHC
 
 | | Crystal structure of cyanovirin-n swapping domain b mutant | | Descriptor: | Cyanovirin-N, PHOSPHATE ION, SODIUM ION | | Authors: | Matei, E, Zheng, A, Furey, W, Rose, J, Aiken, C, Gronenborn, A.M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization.
J.Biol.Chem., 285, 2010
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4WHJ
 
 | |
4J93
 
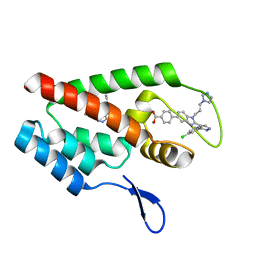 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
5UP4
 
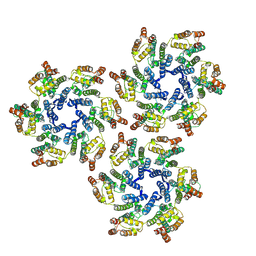 | |
3NTE
 
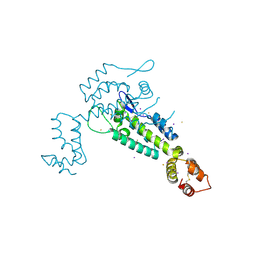 | |
4B3N
 
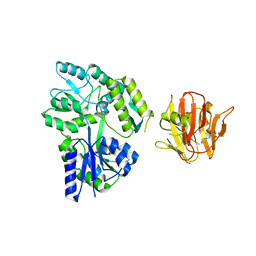 | | Crystal structure of rhesus TRIM5alpha PRY/SPRY domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MALTOSE-BINDING PERIPLASMIC PROTEIN, TRIPARTITE MOTIF-CONTAINING PROTEIN 5, ... | | Authors: | Yang, H, Ji, X, Zhao, Q, Xiong, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insight Into HIV-1 Capsid Recognition by Rhesus Trim5Alpha
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KOD
 
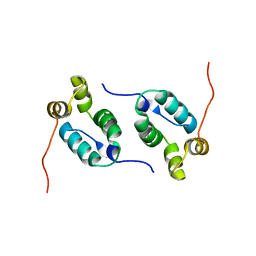 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
4XRO
 
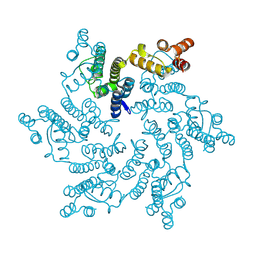 | |
4XRQ
 
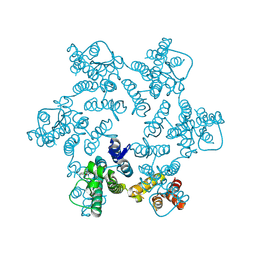 | |
9D6E
 
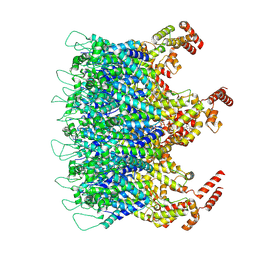 | | Gag CA-SP1 immature lattice bound with Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9D6D
 
 | |
9D88
 
 | |
9D6C
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9DWD
 
 | |
9CWV
 
 | |
7ASL
 
 | |
7ASH
 
 | |
4FBV
 
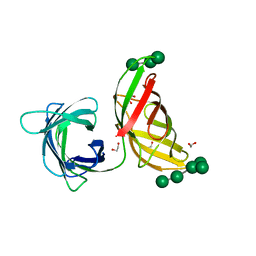 | | Crystal structure of the Myxococcus Xanthus hemagglutinin in complex with a3,a6-mannopentaose | | Descriptor: | 1,2-ETHANEDIOL, Myxobacterial hemagglutinin, alpha-D-mannopyranose, ... | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into the Anti-HIV Activity of the Oscillatoria agardhii Agglutinin Homolog Lectin Family.
J.Biol.Chem., 287, 2012
|
|
4FBR
 
 | |
4FBO
 
 | |
6AYA
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Nup153 peptide | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
