2IMO
 
 | |
2IMG
 
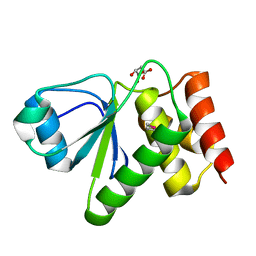 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
4QUQ
 
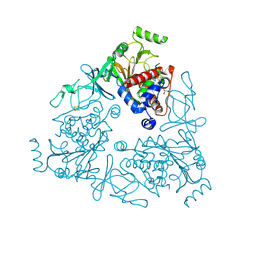 | | Crystal structure of stachydrine demethylase in complex with azide | | Descriptor: | AZIDE ION, COBALT HEXAMMINE(III), FE (III) ION, ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.266 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
4QUP
 
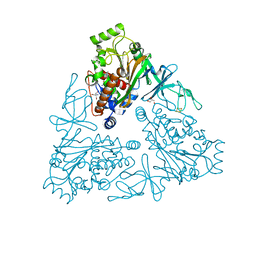 | | Crystal structure of stachydrine demethylase with N-methyl proline from low X-ray dose composite datasets | | Descriptor: | 1-methyl-L-proline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT HEXAMMINE(III), ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
4QUR
 
 | | Crystal Structure of stachydrine demethylase in complex with cyanide, oxygen, and N-methyl proline in a new orientation | | Descriptor: | 1-methyl-L-proline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT HEXAMMINE(III), ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
3D3X
 
 | |
3CMN
 
 | |
3CVG
 
 | |
3D5L
 
 | |
1XD7
 
 | |
1X77
 
 | | Crystal structure of a NAD(P)H-dependent FMN reductase complexed with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, conserved hypothetical protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-08-13 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3KD9
 
 | |
3LXT
 
 | |
3M0F
 
 | |
3LKB
 
 | |
3MQT
 
 | |
3DH0
 
 | |
3DLI
 
 | |
3EHE
 
 | |
3E3V
 
 | |
3FII
 
 | |
3FIE
 
 | |
3HUU
 
 | |
3HS3
 
 | |
4DPO
 
 | | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1 | | Descriptor: | Conserved protein | | Authors: | Agarwal, R, Chamala, S, Evans, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Foti, R, Siedel, R, Zencheck, W, Villigas, G, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-13 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1
To be Published
|
|
