1OM2
 
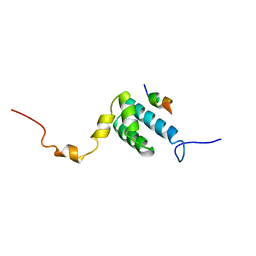 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
3WW6
 
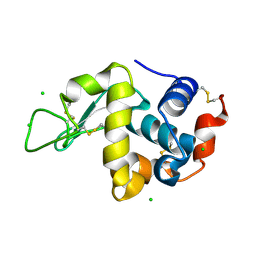 | | Crystal Structure of hen egg white lysozyme mutant N46D/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
3WW5
 
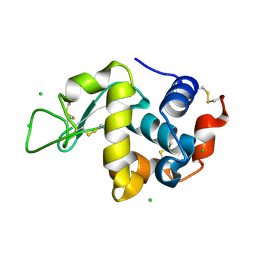 | | Crystal Structure of hen egg white lysozyme mutant N46E/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
5YCQ
 
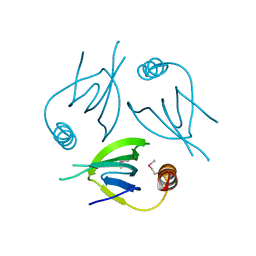 | | Unique Specificity-Enhancing Factor for the AAA+ Lon Protease | | Descriptor: | Heat shock protein HspQ | | Authors: | Abe, Y, Shioi, S, Kita, S, Nakata, H, Maenaka, K, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2017-09-08 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | X-ray crystal structure of Escherichia coli HspQ, a protein involved in the retardation of replication initiation
FEBS Lett., 591, 2017
|
|
2D35
 
 | | Solution structure of Cell Division Reactivation Factor, CedA | | Descriptor: | Cell division activator cedA | | Authors: | Abe, Y, Watanabe, N, Matsuda, Y, Yoshida, Y, Katayama, T, Ueda, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis and Molecular Interaction of Cell Division Reactivation Factor, CedA from Escherichia coli
To be Published
|
|
2KZB
 
 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2E0G
 
 | |
7YPD
 
 | |
2KZK
 
 | | Solution structure of alpha-mannosidase binding domain of Atg34 | | Descriptor: | Uncharacterized protein YOL083W | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
3WZU
 
 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YTV
 
 | | Crystal structure of Mdm35 | | Descriptor: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTW
 
 | | Crystal structure of Ups1-Mdm35 complex | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTX
 
 | | Crystal structure of Ups1-Mdm35 complex with PA | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
5B2L
 
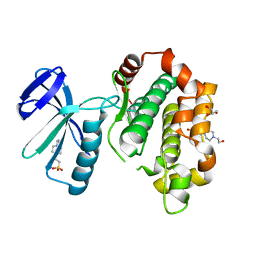 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2M
 
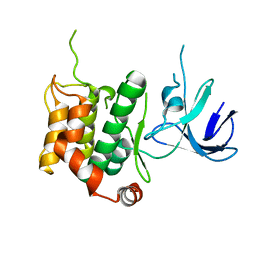 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2K
 
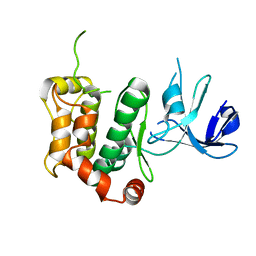 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
6J7C
 
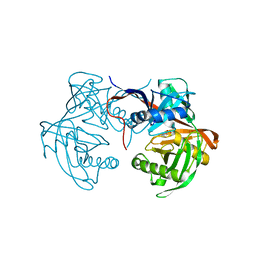 | | Crystal structure of proline racemase-like protein from Thermococcus litoralis in complex with proline | | Descriptor: | PROLINE, Proline racemase | | Authors: | Watanabe, Y, Watanabe, S, Itoh, Y, Watanabe, Y. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of substrate-bound bifunctional proline racemase/hydroxyproline epimerase from a hyperthermophilic archaeon.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
3VU4
 
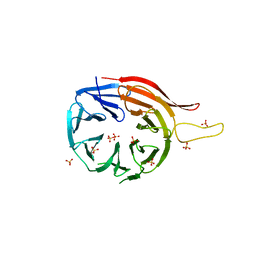 | |
7BYU
 
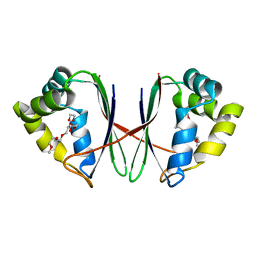 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
7BYW
 
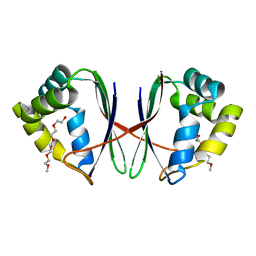 | |
5AUJ
 
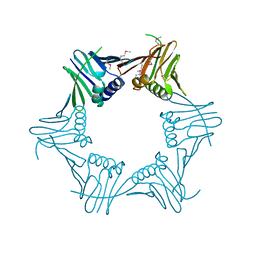 | |
5AXC
 
 | | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin | | Descriptor: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin
To Be Published
|
|
5B0U
 
 | |
5AXD
 
 | |
5AXB
 
 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | Descriptor: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
