7A3H
 
 | | NATIVE ENDOGLUCANASE CEL5A CATALYTIC CORE DOMAIN AT 0.95 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, ETHANOL, GLYCEROL | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-08-05 | | Release date: | 1999-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
4V44
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
4V45
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
1BSI
 
 | | HUMAN PANCREATIC ALPHA-AMYLASE FROM PICHIA PASTORIS, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Sidhu, G, Vo, H.C, Hewitt, J, Cote, H.C.F, Wang, Y, Numao, S, Macgillivray, R.T.A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, mutagenesis, and structural analysis of human pancreatic alpha-amylase expressed in Pichia pastoris.
Protein Sci., 8, 1999
|
|
1C5H
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
1C5I
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6DFE
 
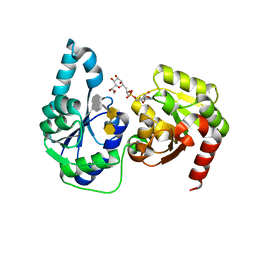 | | The structure of a ternary complex of E. coli WaaC | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-acetamido-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADP-heptose--LPS heptosyltransferase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R)-1-{(2S,3S,4R,5S,6R)-6-[(1S)-1,2-dihydroxyethyl]-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl}propan-2-yl hydrogen (R)-phosphate (non-preferred name) | | Authors: | Worrall, L.J, Blaukopf, M, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Insights into Heptosyltransferase I Catalysis and Inhibition through the Structure of Its Ternary Complex.
Structure, 26, 2018
|
|
2HIS
 
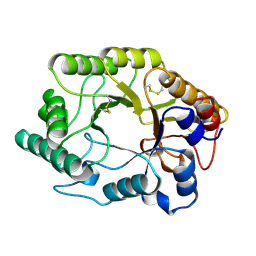 | | CELLULOMONAS FIMI XYLANASE/CELLULASE DOUBLE MUTANT E127A/H205N WITH COVALENT CELLOBIOSE | | Descriptor: | CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Notenboom, V, Birsan, C, Nitz, M, Rose, D.R, Warren, R.A.J, Wither, S.G. | | Deposit date: | 1998-02-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into transition state stabilization of the beta-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants.
Nat.Struct.Biol., 5, 1998
|
|
3IJ8
 
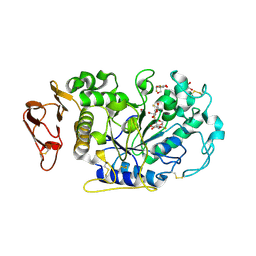 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, 5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ7
 
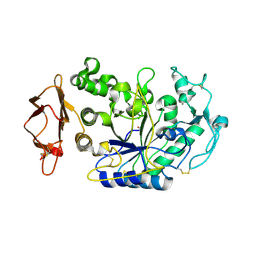 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | 4-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranosyl fluoride, 4-O-methyl-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-2)-5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-03 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ9
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
1EXP
 
 | | BETA-1,4-GLYCANASE CEX-CD | | Descriptor: | BETA-1,4-D-GLYCANASE CEX-CD, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | White, A, Tull, D, Johns, K.L, Withers, S.G, Rose, D.R. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic observation of a covalent catalytic intermediate in a beta-glycosidase.
Nat.Struct.Biol., 3, 1996
|
|
6N1B
 
 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
6N1A
 
 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
6Q1P
 
 | | Glucocerebrosidase in complex with pharmacological chaperone norIMX8 | | Descriptor: | (2S,3S,4S,5R)-2-{[(4-methylpentyl)sulfonyl]methyl}piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
6Q1N
 
 | | Glucocerebrosidase in complex with pharmacological chaperone IMX8 | | Descriptor: | (2R,3S,4S,5R)-2-[2-(methylsulfanyl)ethyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.526 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
3LMY
 
 | | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bateman, K.S, Cherney, M.M, Withers, S.G, Mahuran, D.J, Tropak, M, James, M.N.G. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine
To be Published
|
|
6A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ENDOGLUCANASE, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
4KGJ
 
 | | Crystal structure of human alpha-L-iduronidase complex with 5-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoro-alpha-L-idopyranosyluronic acid fluoride, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KH2
 
 | | Crystal structure of human alpha-L-iduronidase complex with 2-deoxy-2-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2,6-anhydro-5-deoxy-5-fluoro-L-idonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KGL
 
 | | Crystal structure of human alpha-L-iduronidase complex with [2R,3R,4R,5S]-2-carboxy-3,4,5-trihydroxy-piperidine | | Descriptor: | (2R,3R,4R,5S)-3,4,5-trihydroxypiperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
