1U07
 
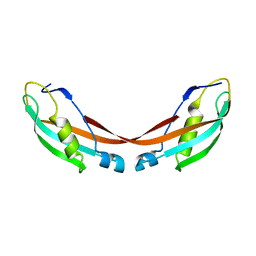 | | Crystal Structure of the 92-residue C-term. part of TonB with significant structural changes compared to shorter fragments | | Descriptor: | TonB protein | | Authors: | Koedding, J, Killig, F, Polzer, P, Howard, S.P, Diederichs, K, Welte, W. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of a 92-residue c-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments
J.Biol.Chem., 280, 2005
|
|
4XXH
 
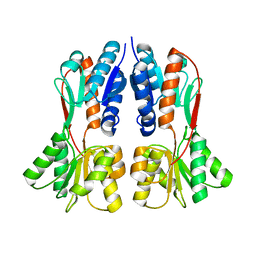 | | TREHALOSE REPRESSOR FROM ESCHERICHIA COLI | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, HTH-type transcriptional regulator TreR | | Authors: | Hars, U, Horlacher, R, Boos, W, Smart, O.S, Bricogne, G, Welte, W, Diederichs, K. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF THE EFFECTOR-BINDING DOMAIN OF THE TREHALOSE-REPRESSOR OF ESCHERICHIA COLI, A MEMBER OF THE LACI FAMILY, IN ITS COMPLEXES WITH INDUCER TREHALOSE-6-PHOSPHATE AND NONINDUCER TREHALOSE.
PROTEIN SCI., 7, 1998
|
|
1FI1
 
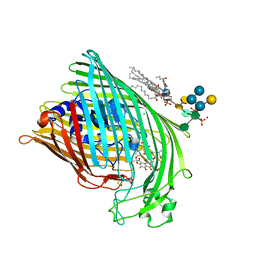 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
1OH2
 
 | |
4CS3
 
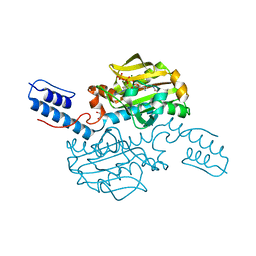 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with an adenylated furan-bearing noncanonical amino acid and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(furan-2-yl)ethyl hydrogen carbonate, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4DFM
 
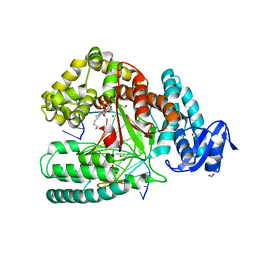 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in ternary complex with 5-(aminopentinyl)-2-dCTP | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(AAAGCGCGCCGTGGTC)-3', 5'-d(GACCACGGCGC ddG)-3', ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
2F5T
 
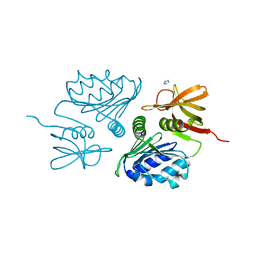 | | Crystal Structure of the sugar binding domain of the archaeal transcriptional regulator TrmB | | Descriptor: | IMIDAZOLE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, archaeal transcriptional regulator TrmB | | Authors: | Krug, M, Lee, S.J, Diederichs, K, Boos, W, Welte, W. | | Deposit date: | 2005-11-27 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Sugar Binding Domain of the Archaeal Transcriptional Regulator TrmB
J.Biol.Chem., 281, 2006
|
|
2FCP
 
 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-15 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
1Z6R
 
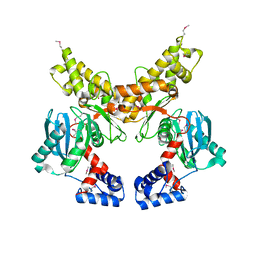 | | Crystal structure of Mlc from Escherichia coli | | Descriptor: | Mlc protein, ZINC ION | | Authors: | Schiefner, A, Gerber, K, Seitz, S, Welte, W, Diederichs, K, Boos, W. | | Deposit date: | 2005-03-23 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli
J.Biol.Chem., 280, 2005
|
|
1A0S
 
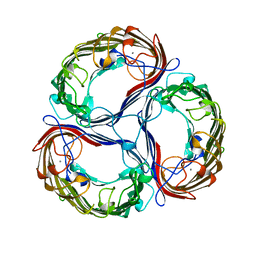 | | SUCROSE-SPECIFIC PORIN | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
1A0T
 
 | | SUCROSE-SPECIFIC PORIN, WITH BOUND SUCROSE MOLECULES | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
4K8Z
 
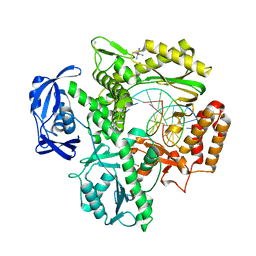 | | KOD Polymerase in binary complex with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, COBALT HEXAMMINE(III), DNA (5'-D(*AP*AP*AP*TP*TP*CP*GP*CP*AP*GP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Bergen, K, Betz, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of KOD and 9N DNA Polymerases Complexed with Primer Template Duplex
Chembiochem, 14, 2013
|
|
2X52
 
 | | CRYSTAL STRUCTURE OF WHEAT GERM AGGLUTININ ISOLECTIN 3 IN COMPLEX WITH A SYNTHETIC DIVALENT CARBOHYDRATE LIGAND | | Descriptor: | AGGLUTININ ISOLECTIN 3, BIS-(2-ACETAMIDO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYLOXYCARBONYL)-4,7,10-TRIOXA-1,13-TRIDECANEDIAMINE, GLYCEROL | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
2X3T
 
 | | Glutaraldehyde-crosslinked wheat germ agglutinin isolectin 1 crystal soaked with a synthetic glycopeptide | | Descriptor: | 2-acetamido-1-O-carbamoyl-2-deoxy-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
2OM5
 
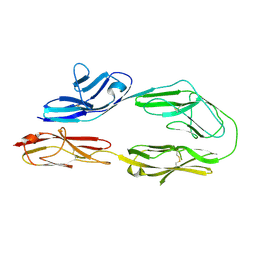 | |
2B4L
 
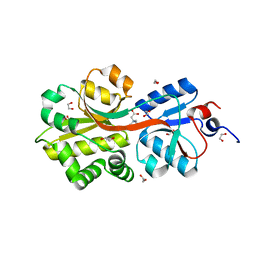 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
1FUE
 
 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
2B4M
 
 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
1KFQ
 
 | | Crystal Structure of Exocytosis-Sensitive Phosphoprotein, pp63/parafusin (Phosphoglucomutse) from Paramecium. OPEN FORM | | Descriptor: | CALCIUM ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-22 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
2UVO
 
 | | High Resolution Crystal Structure of Wheat Germ Agglutinin in Complex with N-Acetyl-D-Glucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN 1, ... | | Authors: | Schwefel, D, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
1URS
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1YCE
 
 | | Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus | | Descriptor: | NONAN-1-OL, SODIUM ION, subunit c | | Authors: | Meier, T, Polzer, P, Diederichs, K, Welte, W, Dimroth, P. | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus.
Science, 308, 2005
|
|
1URD
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1URG
 
 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
