1HNW
 
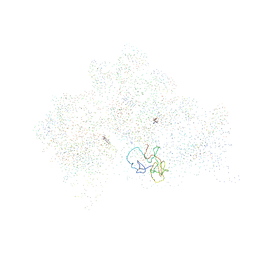 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HR0
 
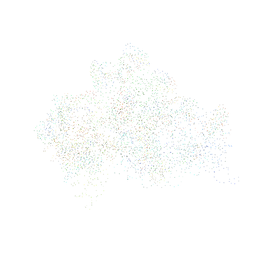 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
5FD9
 
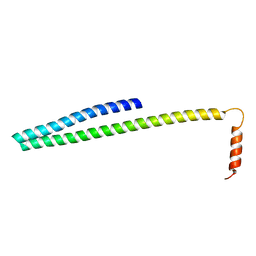 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation B) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
1QB2
 
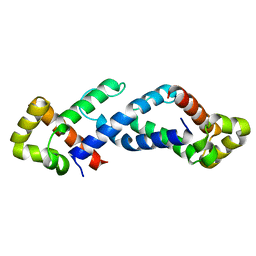 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
4PWX
 
 | |
2AX4
 
 | | Crystal structure of the kinase domain of human 3'-phosphoadenosine 5'-phosphosulphate synthetase 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 | | Authors: | Rabeh, W.M, Nedyalkova, L, Ismail, S, Park, H, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-02 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the kinase domain of PAPSS 2
To be Published
|
|
3LKU
 
 | |
2KZH
 
 | |
2XCN
 
 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | MAGNESIUM ION, N-[(CYCLOPENTYLOXY)CARBONYL]-3-METHYL-L-VALYL-(4R)-N-{(1R)-3-HYDROXY-1-[HYDROXY(OXIDO)BORANYL]PROPYL}-4-(ISOQUINOLIN-1-YLOXY)-L-PROLINAMIDE, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCF
 
 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | CYCLOPENTYL N-[(2S)-1-[(2S,4R)-2-[[(4R)-8-HYDROXY-1,6,10-TRIOXA-5$L^{4}-BORASPIRO[4.5]DECAN-4-YL]CARBAMOYL]-4-ISOQUINOLIN-1-YLOXY-PYRROLIDIN-1-YL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XNI
 
 | | Protein-ligand complex of a novel macrocyclic HCV NS3 protease inhibitor derived from amino cyclic boronates | | Descriptor: | (1-{[(10-tert-butyl-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19,23,23a-tetradecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosin-7(3H)-yl)carbonyl]amino}-3-hydroxypropyl)(trihydroxy)borate(1-), MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Zhou, Y, Li, Q, Plattner, J.J, Baker, S.J, Zhang, S, Kazmierski, W.M, Wright, L.L, Smith, G.K, Grimes, R.M, Crosby, R.M, Creech, K.L, Carballo, L.H, Slater, M.J, Jarvest, R.L, Thommes, P, Hubbard, J.A, Convery, M.A, Nassau, P.M, McDowell, W, Skarzynski, T.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, Pennicott, L.E, Zou, W, Wright, J. | | Deposit date: | 2010-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Macrocyclic Hcv Ns3 Protease Inhibitors Derived from Alpha-Amino Cyclic Boronates.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5NRF
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7i | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-chlorophenyl)ethyl]-~{N}-(phenylmethyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
2LO0
 
 | |
4WWR
 
 | |
1FJG
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
6Q1W
 
 | |
6Q22
 
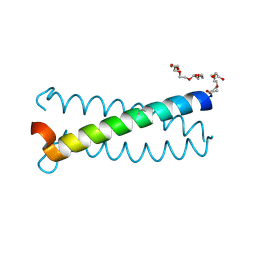 | | Coiled-coil Trimer with Glu:Aminobutyric acid:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:Aminobutyric acid:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-08-06 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6Q25
 
 | |
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1BRV
 
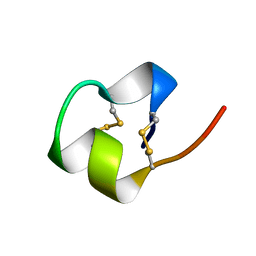 | | SOLUTION NMR STRUCTURE OF THE IMMUNODOMINANT REGION OF PROTEIN G OF BOVINE RESPIRATORY SYNCYTIAL VIRUS, 48 STRUCTURES | | Descriptor: | PROTEIN G | | Authors: | Doreleijers, J.F, Langedijk, J.P.M, Hard, K, Rullmann, J.A.C, Boelens, R, Schaaper, W.M, Van Oirschot, J.T, Kaptein, R. | | Deposit date: | 1996-03-29 | | Release date: | 1997-06-05 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus.
Biochemistry, 35, 1996
|
|
8QAC
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil 8-helix bundle with 4 heptad repeats, antiparallel 8-helix bundle-GLIA | | Descriptor: | antiparallel 8-helix bundle-GLIA | | Authors: | Albanese, K.I, Dawson, W.M, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
5OOB
 
 | |
