1XEB
 
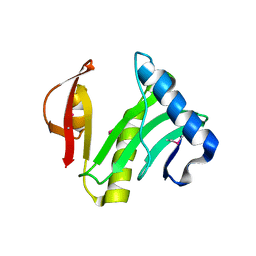 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA0115 | | Authors: | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
2ZI8
 
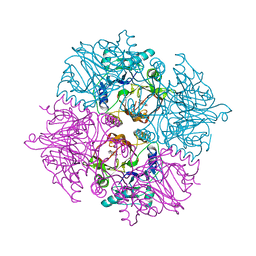 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
3AFF
 
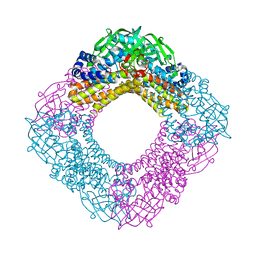 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
2ZYQ
 
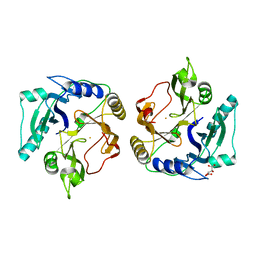 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis | | Descriptor: | D(-)-TARTARIC ACID, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2009-01-28 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
3AFE
 
 | | Crystal structure of the HsaA monooxygenase from M.tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Eltis, L.D, Strynadka, N. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
1JTG
 
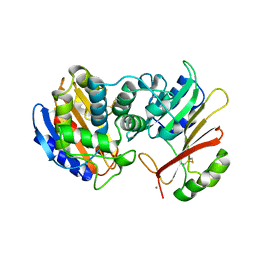 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
3SPU
 
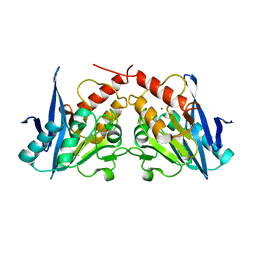 | | apo NDM-1 Crystal Structure | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2011-07-03 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance
Protein Sci., 20, 2011
|
|
2MKY
 
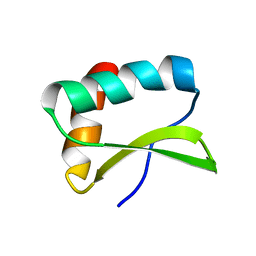 | |
2ZYL
 
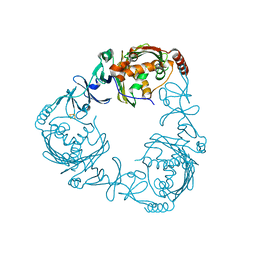 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, POSSIBLE OXIDOREDUCTASE, ... | | Authors: | D'Angelo, I, Capyk, J, Strynadka, N, Eltis, L. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis
J.Biol.Chem., 284, 2009
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
