2GGF
 
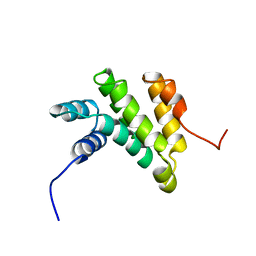 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | Descriptor: | Programmed cell death 4, isoform 1 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
1IW7
 
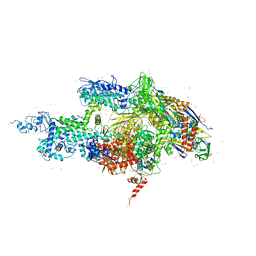 | |
5WVU
 
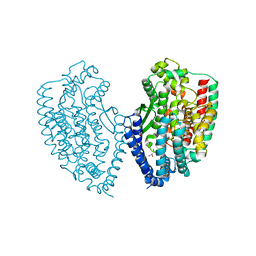 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
3MCJ
 
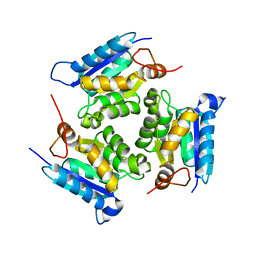 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3CQ3
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8
To be Published
|
|
3CQ1
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8
To be Published
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7CKJ
 
 | | Crystal structure of CMP kinase in complex with CMP from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of Thermus thermophilus CMP kinase complexed with a phosphoryl group acceptor and donor.
PLoS ONE, 15, 2020
|
|
1NZA
 
 | | Divalent cation tolerance protein (Cut A1) from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Divalent cation tolerance protein, GLYCEROL, ... | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1NZ8
 
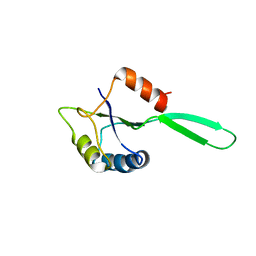 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1N77
 
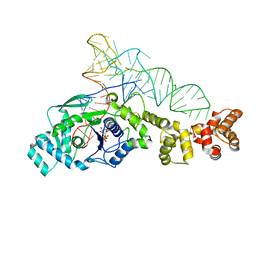 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
3VQX
 
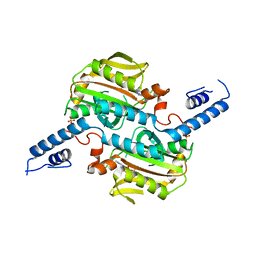 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in triclinic crystal form | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Pyrrolysine--tRNA ligase, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQW
 
 | | Crystal structure of the SeMet substituted catalytic domain of pyrrolysyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VNP
 
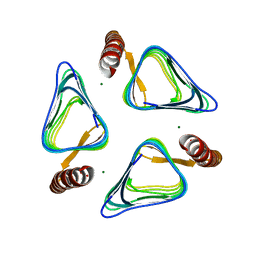 | | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus | | Descriptor: | ACETIC ACID, Hypothetical conserved protein, MAGNESIUM ION | | Authors: | Karthe, P.P, Kumarevel, T.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus
To be Published
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
3VIU
 
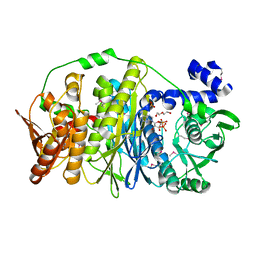 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7FBR
 
 | |
7FBV
 
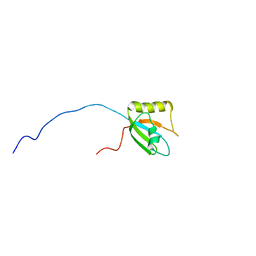 | |
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
2RBG
 
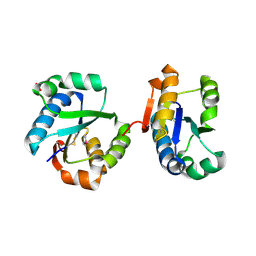 | |
2RQR
 
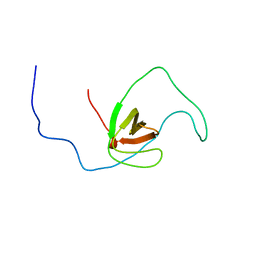 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | Descriptor: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | Authors: | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2RRA
 
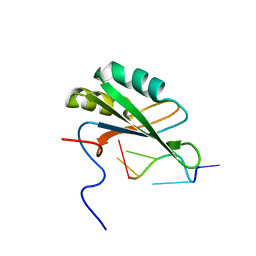 | | Solution structure of RNA binding domain in human Tra2 beta protein in complex with RNA (GAAGAA) | | Descriptor: | 5'-R(*GP*AP*AP*GP*AP*A)-3', cDNA FLJ40872 fis, clone TUTER2000283, ... | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
