6OF3
 
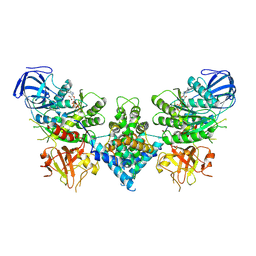 | | Precursor ribosomal RNA processing complex, State 1. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF2
 
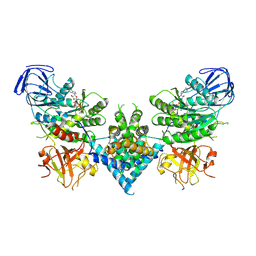 | | Precursor ribosomal RNA processing complex, State 2. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF4
 
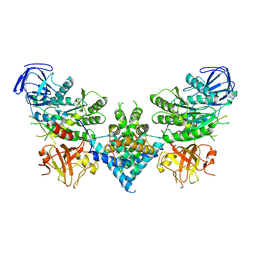 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7K0R
 
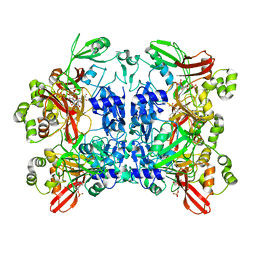 | | Nucleotide bound SARS-CoV-2 Nsp15 | | Descriptor: | PHOSPHATE ION, URIDINE-5'-MONOPHOSPHATE, Uridylate-specific endoribonuclease | | Authors: | Pillon, M.C, Stanley, R.E. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.
Nat Commun, 12, 2021
|
|
3KDK
 
 | |
3KDG
 
 | |
3GAB
 
 | | C-terminal domain of Bacillus subtilis MutL crystal form I | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Guarne, A, Pillon, M.C, Lorenowicz, J.J, Mitchell, R.R, Chung, Y.S, Friedhoff, P. | | Deposit date: | 2009-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol.Cell, 39, 2010
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5JMV
 
 | | Crystal structure of mjKae1-pfuPcc1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, ... | | Authors: | Wan, L, Sicheri, F. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3864696 Å) | | Cite: | Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis.
Nucleic Acids Res., 44, 2016
|
|
7N06
 
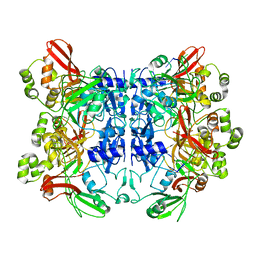 | | SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state | | Descriptor: | RNA (5'-R(*AP*UP*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
7N33
 
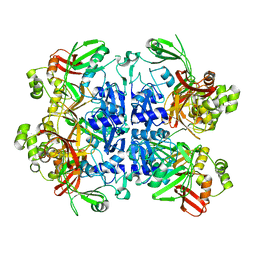 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
6E8E
 
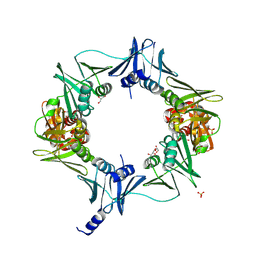 | |
6E8D
 
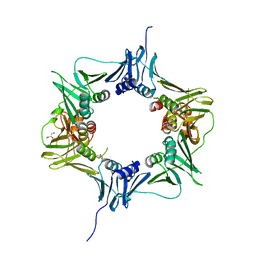 | |
4O1O
 
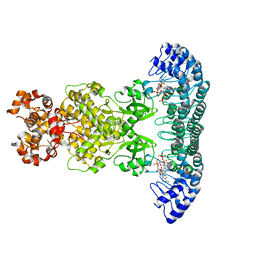 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4O1P
 
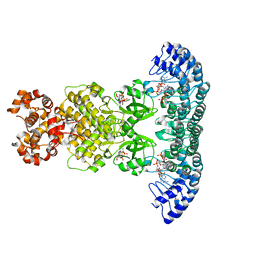 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
5CW6
 
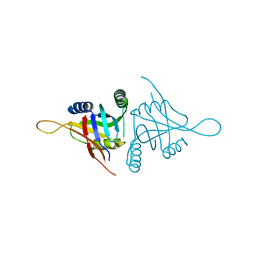 | | Structure of metal dependent enzyme DrBRCC36 | | Descriptor: | DrBRCC36, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.193 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW4
 
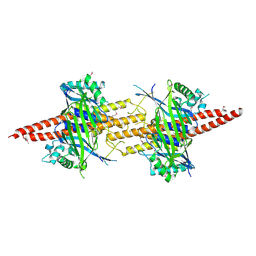 | | Structure of CfBRCC36-CfKIAA0157 complex (Selenium Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, GLYCEROL, Protein FAM175B, ... | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW5
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (QSQ mutant) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.736 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW3
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Zn Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5FBT
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FBS
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with ADP and magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoenolpyruvate synthase | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
