2ESK
 
 | |
2ESQ
 
 | |
2ESO
 
 | |
2ESP
 
 | | Human ubiquitin-conjugating enzyme (E2) UbcH5b mutant Ile88Ala | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Ozkan, E, Yu, H, Deisenhofer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4OFD
 
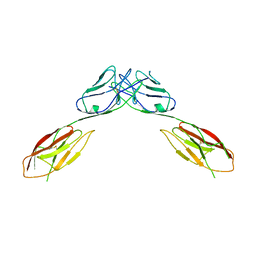 | | Crystal Structure of mouse Neph1 D1-D2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of IRRE-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF3
 
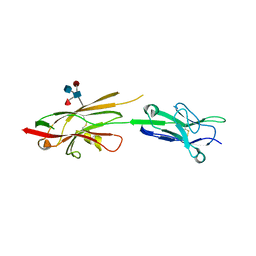 | | Crystal Structure of SYG-1 D1-D2, Glycosylated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFP
 
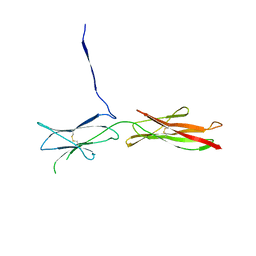 | | Crystal Structure of SYG-2 D3-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-2 | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF8
 
 | | Crystal Structure of Rst D1-D2 | | Descriptor: | GLYCEROL, Irregular chiasm C-roughest protein, SODIUM ION | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF7
 
 | | Crystal Structure of SYG-1 D1, Crystal Form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFY
 
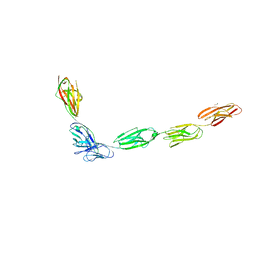 | | Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHYL MERCURY ION, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFI
 
 | | Crystal Structure of Duf (Kirre) D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of irre, isoform A, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF0
 
 | | Crystal Structure of SYG-1 D1-D2, refolded | | Descriptor: | Protein SYG-1, isoform b | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFK
 
 | | Crystal Structure of SYG-2 D4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ozkan, E, Borek, D, Otwinowski, Z, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF6
 
 | | Crystal Structure of SYG-1 D1, Crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
5EO9
 
 | | Crystal Structure of the complex of Dpr6 Domain 1 bound to DIP-alpha Domain 1+2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CG32791, isoform A, ... | | Authors: | Ozkan, E, Zinn, K, Garcia, K.C. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2988 Å) | | Cite: | Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins.
Cell, 163, 2015
|
|
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6PLL
 
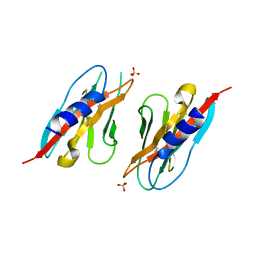 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6POL
 
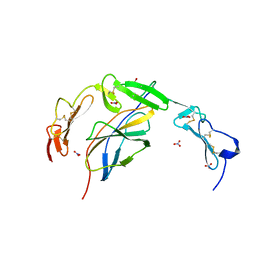 | | Crystal structure of the human NELL1 EGF1-3-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
8TK9
 
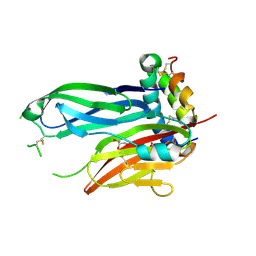 | | ZIG-4-INS-6 complex, tetragonal form | | Descriptor: | GLYCEROL, Probable insulin-like peptide beta-type 5, SULFATE ION, ... | | Authors: | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways
To Be Published
|
|
8TKU
 
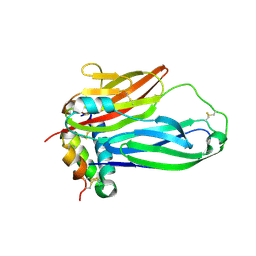 | | ZIG-4-INS-6 complex, primitive monoclinic form | | Descriptor: | Probable insulin-like peptide beta-type 5, Zwei Ig domain protein zig-4 | | Authors: | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways
To Be Published
|
|
8TKT
 
 | | ZIG-4-INS-6 complex, C-centered monoclinic form | | Descriptor: | Probable insulin-like peptide beta-type 5, Zwei Ig domain protein zig-4 | | Authors: | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways
To Be Published
|
|
8SUF
 
 | | The complex of TOL-1 ectodomain bound to LAT-1 Lectin domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Latrophilin-like protein 1, ... | | Authors: | Carmona Rosas, G, Li, J, Arac, D, Ozkan, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis and functional roles for Toll-like receptor binding to Latrophilin adhesion-GPCR in embryo development
To Be Published
|
|
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
3BIX
 
 | | Crystal structure of the extracellular esterase domain of Neuroligin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
8EDC
 
 | | Structure of C. elegans UNC-5 IG 1+2 Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Netrin receptor unc-5, SULFATE ION | | Authors: | Priest, J.M, Ozkan, E. | | Deposit date: | 2022-09-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv, 10, 2024
|
|
