1AV5
 
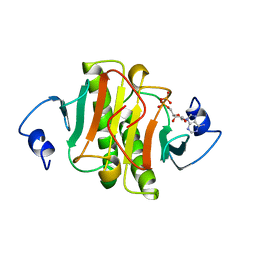 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
6FIT
 
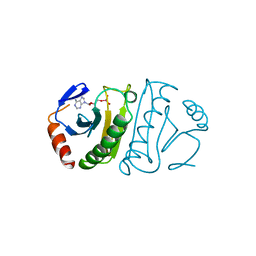 | | FHIT-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE MONOTUNGSTATE, FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5FIT
 
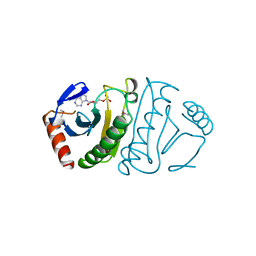 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1KPE
 
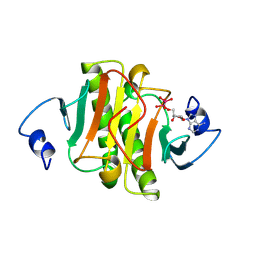 | | PKCI-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE-5'-DITUNGSTATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1KPF
 
 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
3EAY
 
 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
3FIT
 
 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) IN COMPLEX WITH ADENOSINE/SULFATE AMP ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRAGILE HISTIDINE PROTEIN, SULFATE ION, ... | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
1JF9
 
 | |
1KMK
 
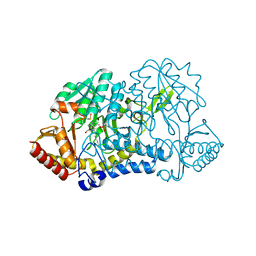 | |
1KMJ
 
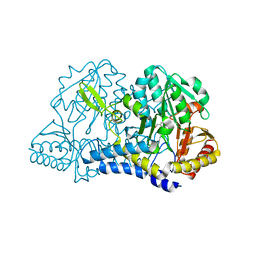 | |
1KPB
 
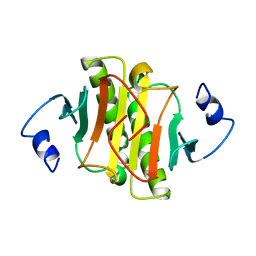 | | PKCI-1-APO | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPA
 
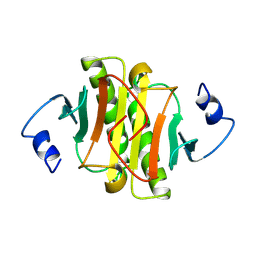 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
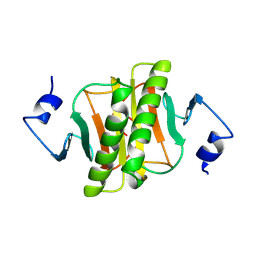 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1ECL
 
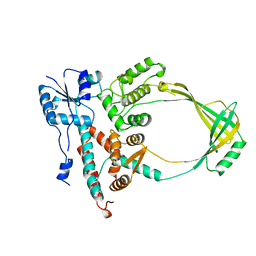 | |
4FIT
 
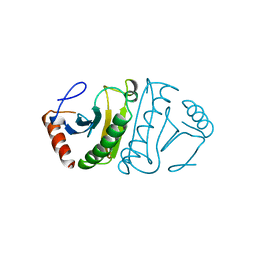 | | FHIT-APO | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1D8H
 
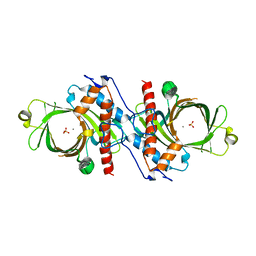 | | X-RAY CRYSTAL STRUCTURE OF YEAST RNA TRIPHOSPHATASE IN COMPLEX WITH SULFATE AND MANGANESE IONS. | | Descriptor: | MANGANESE (II) ION, SULFATE ION, mRNA TRIPHOSPHATASE CET1 | | Authors: | Lima, C.D, Wang, L.K, Shuman, S. | | Deposit date: | 1999-10-24 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus.
Cell(Cambridge,Mass.), 99, 1999
|
|
1D8I
 
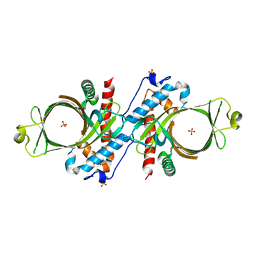 | |
1FIT
 
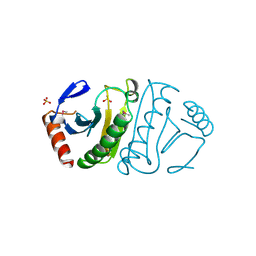 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
1RI6
 
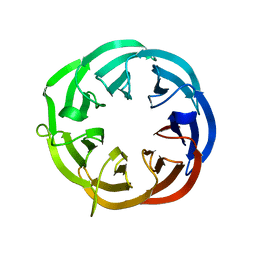 | | Structure of a putative isomerase from E. coli | | Descriptor: | putative isomerase ybhE | | Authors: | Lima, C.D, Kniewel, R, Solorzano, V, Wu, J, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative 7-bladed propeller isomerase
To be Published
|
|
2FIT
 
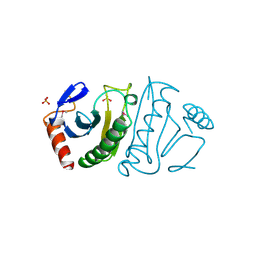 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
4M52
 
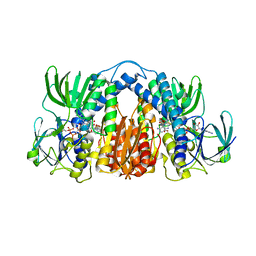 | | Structure of Mtb Lpd bound to SL827 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-[(2-amino-5-bromopyridin-3-yl)sulfonyl]-N-(4-methoxyphenyl)-N~2~-methylglycinamide | | Authors: | Lima, C.D. | | Deposit date: | 2013-08-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase.
Biochemistry, 52, 2013
|
|
3KYD
 
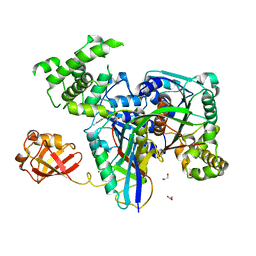 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
6DG4
 
 | |
3KYC
 
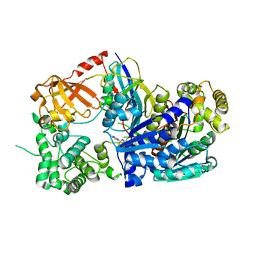 | | Human SUMO E1 complex with a SUMO1-AMP mimic | | Descriptor: | 5'-deoxy-5'-(sulfamoylamino)adenosine, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
3KYH
 
 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
