3LOB
 
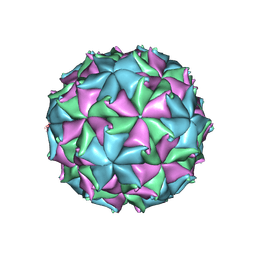 | | Crystal Structure of Flock House Virus calcium mutant | | Descriptor: | Coat protein beta, Coat protein gamma, RNA (5'-R(*UP*UP*U*AP*UP*CP*UP*(P))-3'), ... | | Authors: | Johnson, J.E, Banerjee, M, Speir, J.A, Huang, R. | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate.
J.Virol., 84, 2010
|
|
1A6C
 
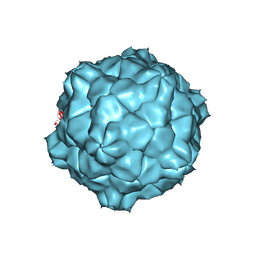 | | STRUCTURE OF TOBACCO RINGSPOT VIRUS | | Descriptor: | TOBACCO RINGSPOT VIRUS CAPSID PROTEIN | | Authors: | Johnson, J.E, Chandrasekar, V. | | Deposit date: | 1998-02-23 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily.
Structure, 6, 1998
|
|
4FRN
 
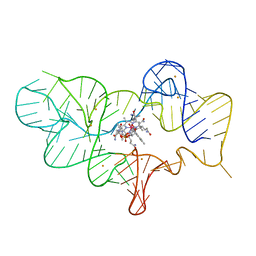 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | Descriptor: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4GMA
 
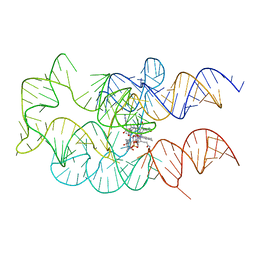 | | Crystal structure of the adenosylcobalamin riboswitch | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
7ANM
 
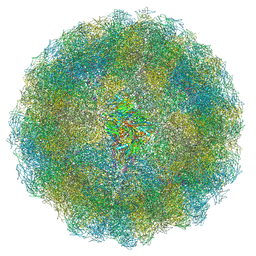 | | Nudaurelia capensis omega virus capsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
7ATA
 
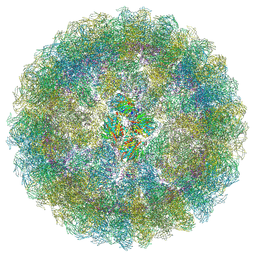 | | Nudaurelia capensis omega virus procapsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.63 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
3E8K
 
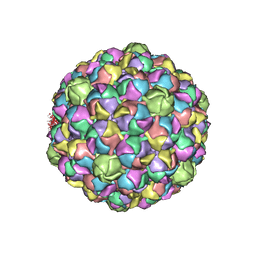 | |
3DDX
 
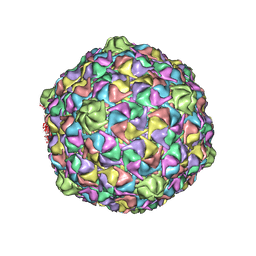 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
3F2E
 
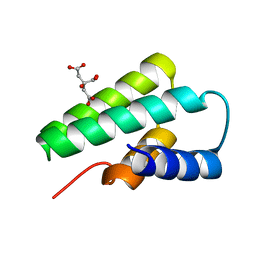 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
3S6P
 
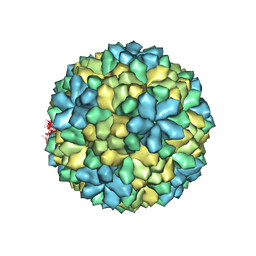 | |
8A41
 
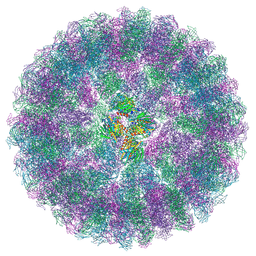 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
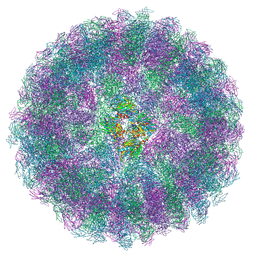 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
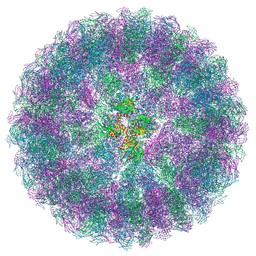 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AC6
 
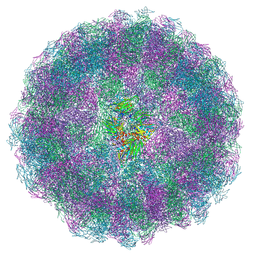 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8ACH
 
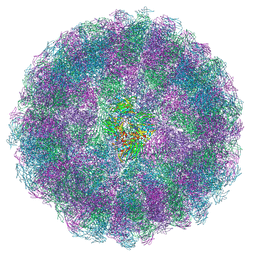 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AAY
 
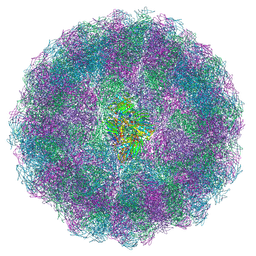 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): small class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
5FMO
 
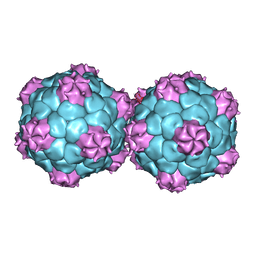 | | Crystal structure and proteomics analysis of empty virus like particles of Cowpea mosaic virus | | Descriptor: | EMPTY VIRUS LIKE PARTICLES OF COWPEA MOSAIC VIRUS | | Authors: | Huynh, N, Hesketh, E.L, Saxena, P, Meshcheriakova, Y, Ku, Y.C, Hoang, L, Johnson, J.E, Ranson, N.A, Lomonossoff, G.P, Reddy, V.S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Proteomics Analysis of Empty Virus-Like Particles of Cowpea Mosaic Virus.
Structure, 24, 2016
|
|
4FSJ
 
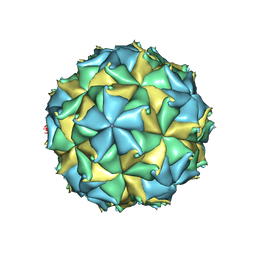 | | Crystal structure of the virus like particle of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FTS
 
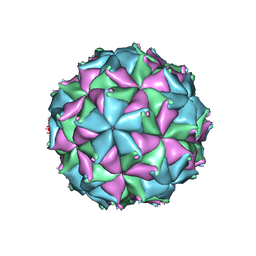 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
1B35
 
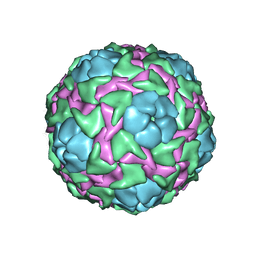 | | CRICKET PARALYSIS VIRUS (CRPV) | | Descriptor: | PROTEIN (CRICKET PARALYSIS VIRUS, VP1), VP2), ... | | Authors: | Tate, J.G, Liljas, L, Scotti, P.D, Christian, P.D, Lin, T.W, Johnson, J.E. | | Deposit date: | 1998-12-17 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cricket paralysis virus: the first view of a new virus family.
Nat.Struct.Biol., 6, 1999
|
|
1BMV
 
 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
4FTE
 
 | | Crystal structure of the D75N mutant capsid of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FTB
 
 | | Crystal structure of the authentic Flock House virus particle | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
3QPR
 
 | | HK97 Prohead I encapsidating inactive virally encoded protease | | Descriptor: | Major capsid protein | | Authors: | Huang, R.K, Khayat, R, Lee, K.K, Gertsman, I, Duda, R.L, Hendrix, R.W, Johnson, J.E. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation.
J.Mol.Biol., 408, 2011
|
|
