4XHS
 
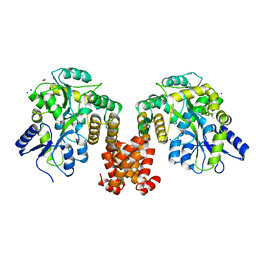 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | Authors: | Jin, T, Huang, M, Jiang, J, Xiao, T. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
4IFP
 
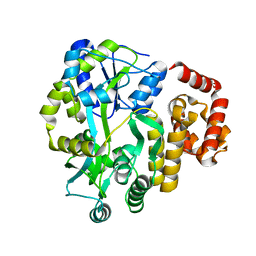 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
4IKM
 
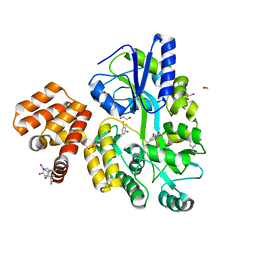 | | X-ray structure of CARD8 CARD domain | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Jin, T, Huang, M, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4606 Å) | | Cite: | The structure of the CARD8 caspase-recruitment domain suggests its association with the FIIND domain and procaspases through adjacent surfaces.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IRL
 
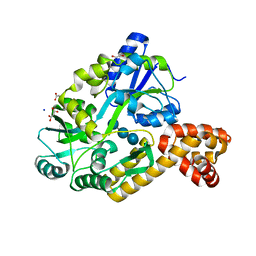 | | X-ray structure of the CARD domain of zebrafish GBP-NLRP1 like protein | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jin, T, Huang, M, Smith, P, Xiao, T. | | Deposit date: | 2013-01-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5WPW
 
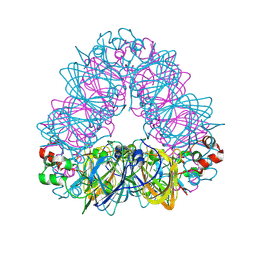 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
3C3V
 
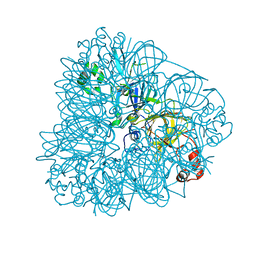 | |
5Y3S
 
 | |
5GPP
 
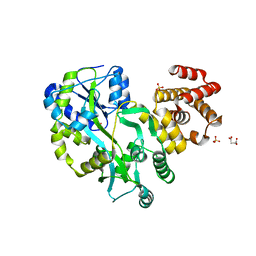 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5GPQ
 
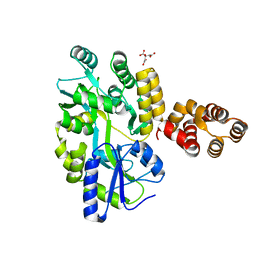 | | Crystal Structure of zebrafish ASC CARD Domain | | Descriptor: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
5Y37
 
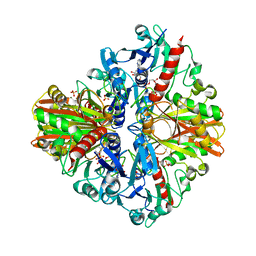 | | Crystal structure of GBS GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Jin, T, Zhou, K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High-resolution crystal structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5Y2G
 
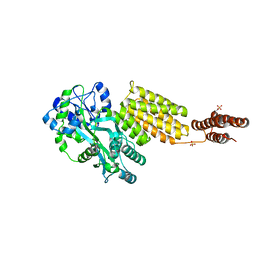 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5YZP
 
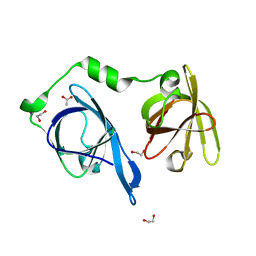 | | Crystal structure of p204 HINa domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ifi204 | | Authors: | Jin, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5Z7D
 
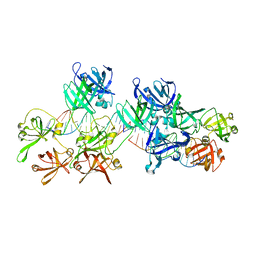 | | p204HINab-dsDNA complex structure | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*CP*AP*GP*AP*AP*AP*GP*AP*GP*AP*GP*C)-3'), Interferon-activable protein 204 | | Authors: | Jin, T, Jiang, J, Xiao, T.S. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5YZW
 
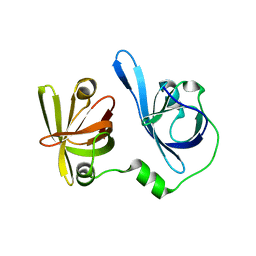 | | Crystal structure of p204 HINb domain | | Descriptor: | Ifi204 | | Authors: | Jin, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5ZNZ
 
 | | Structure of mDR3 DD with MBP tag mutant-I387V | | Descriptor: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | Authors: | Jin, T, Yin, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5K8I
 
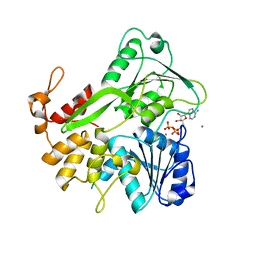 | | Crystal structure of ZIKV NS3 helicase in complex with ATP and Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8T
 
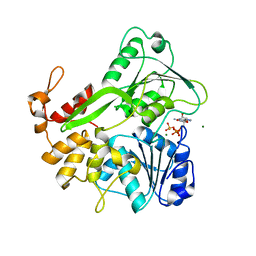 | | Crystal structure of ZIKV NS3 helicase in complex with GTP-gammar S and an magnesium ion | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5GP1
 
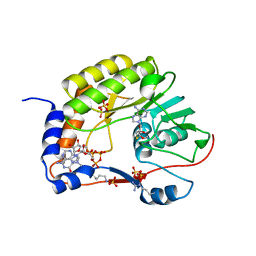 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5GOZ
 
 | |
5JWH
 
 | | Apo structure | | Descriptor: | 1,2-ETHANEDIOL, NS3 helicase | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8L
 
 | | Crystal structure of ZIKV NS3 helicase in complex with GTP-gammar S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8U
 
 | | Crystal structure of ZIKV NS3 helicase in complex with ADP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
7FCQ
 
 | |
