3ZIF
 
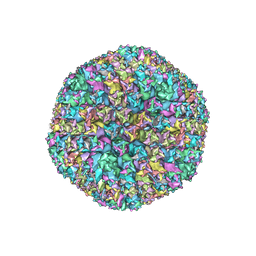 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
3J17
 
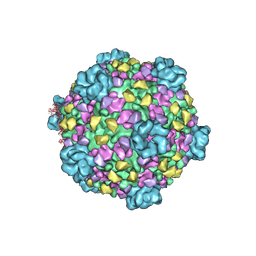 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IK3
 
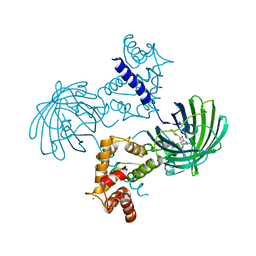 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK9
 
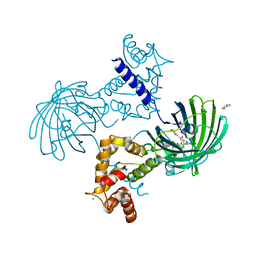 | | High resolution structure of GCaMP3 dimer form 2 at pH 7.5 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, RCaMP, ... | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK8
 
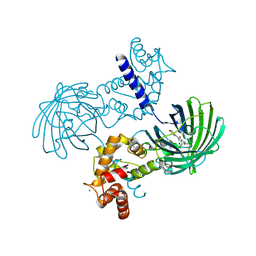 | | High resolution structure of GCaMP3 dimer form 1 at pH 7.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK5
 
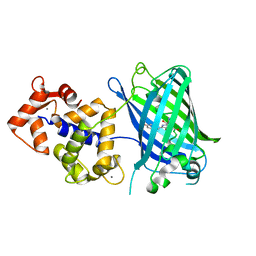 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK1
 
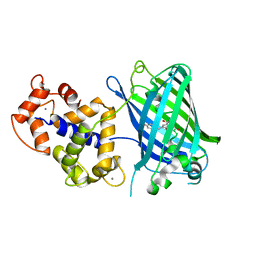 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
3JB8
 
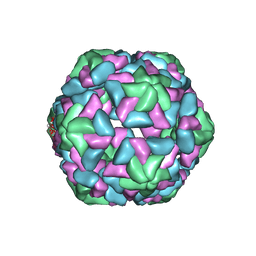 | | Insight into Three-dimensional structure of Maize Chlorotic Mottle Virus Revealed by Single Particle Analysis | | Descriptor: | Coat protein | | Authors: | Wang, C.Y, Zhang, Q.F, Gao, Y.Z, Zhou, X.P, Ji, G, Huang, X.J, Hong, J, Zhang, C.X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Insight into the three-dimensional structure of maize chlorotic mottle virus revealed by Cryo-EM single particle analysis.
Virology, 485, 2015
|
|
3J6Q
 
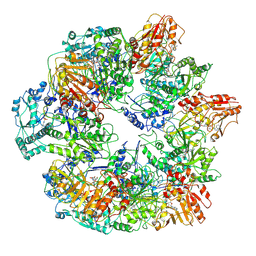 | | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Structural protein VP3 | | Authors: | Zhu, B, Yang, C, Liu, H, Cheng, L, Song, F, Zeng, S, Huang, X, Ji, G, Zhu, P. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus.
J.Mol.Biol., 426, 2014
|
|
3IZ3
 
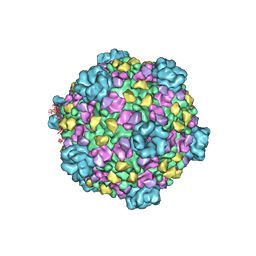 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6M54
 
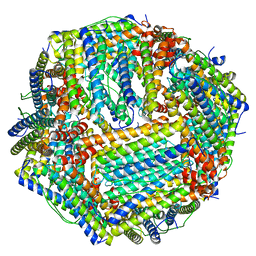 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
6M52
 
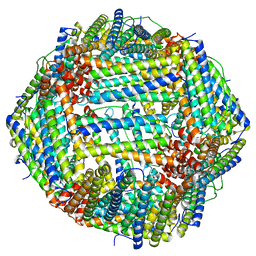 | | Human apo ferritin frozen on TEM grid with amorphous carbon supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
8XQ4
 
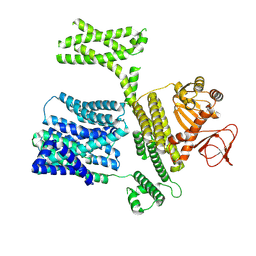 | |
8XQ8
 
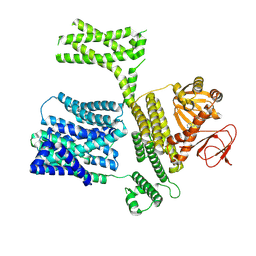 | |
8XQA
 
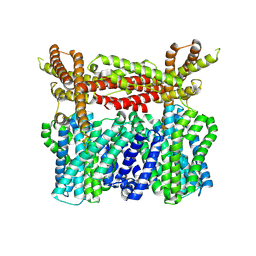 | |
8XQ7
 
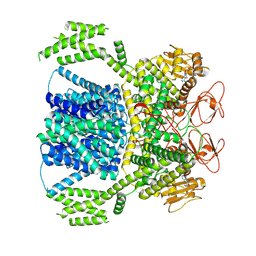 | |
8XPQ
 
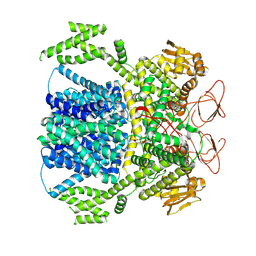 | |
8XQ9
 
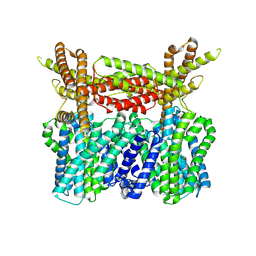 | |
3J1E
 
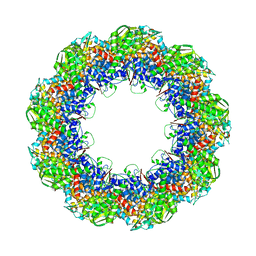 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in apo state | | Descriptor: | Chaperonin beta subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1B
 
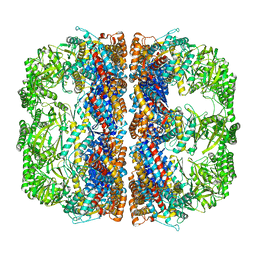 | | Cryo-EM structure of 8-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1C
 
 | | Cryo-EM structure of 9-fold symmetric rATcpn-alpha in apo state | | Descriptor: | Chaperonin alpha subunit | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
3J1F
 
 | | Cryo-EM structure of 9-fold symmetric rATcpn-beta in ATP-binding state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin beta subunit, MAGNESIUM ION | | Authors: | Zhang, K, Wang, L, Liu, Y.X, Wang, X, Gao, B, Hu, Z.J, Ji, G, Chan, K.Y, Schulten, K, Dong, Z.Y, Sun, F. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Flexible interwoven termini determine the thermal stability of thermosomes.
Protein Cell, 4, 2013
|
|
