4PQE
 
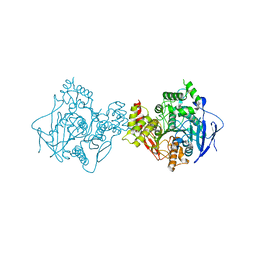 | | Crystal Structure of Human Acetylcholinesterase | | Descriptor: | Acetylcholinesterase | | Authors: | Dym, O, Unger, T, Toker, L, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-03-02 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Acetylcholinesterase
To be Published
|
|
3KIH
 
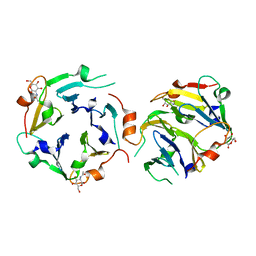 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KIF
 
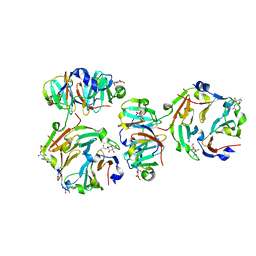 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib1-B7-18 and Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin, SULFATE ION | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4FL0
 
 | | Crystal structure of ALD1 from Arabidopsis thaliana | | Descriptor: | Aminotransferase ALD1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sobolev, V, Edelman, M, Dym, O, Unger, T, Albeck, S, Kirma, M, Galili, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2012-06-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ALD1, a plant-specific homologue of the universal diaminopimelate aminotransferase enzyme of lysine biosynthesis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4QXU
 
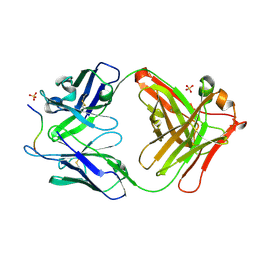 | | Novel Inhibition Mechanism of Membrane Metalloprotease by an Exosite-Swiveling Conformational antibody | | Descriptor: | Matrix metalloproteinase-14, SULFATE ION, anti_MT1-MMP Heavy chain, ... | | Authors: | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Moreno, v, Cuiniasse, P, Dive, V, Arroyo, A.G, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-07-22 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
2NVB
 
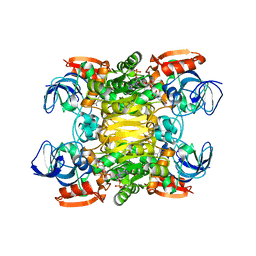 | | Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases (ADHs) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Goihberg, E, Tel-Or, S, Peretz, M, Frolow, F, Dym, O, Burstein, Y, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution
Proteins, 72, 2008
|
|
3D3M
 
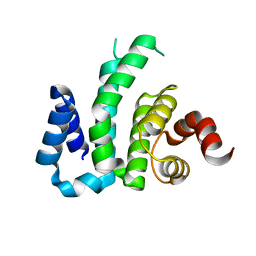 | |
4OUU
 
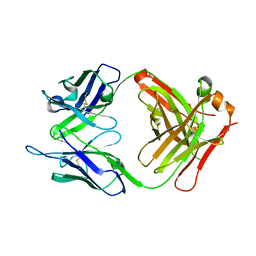 | | anti-MT1-MMP monoclonal antibody | | Descriptor: | anti_MT1-MMP Heavy chain, anti_MT1-MMP light chain | | Authors: | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Koziol, A, Cuniasse, P, Dive, V, Arroyo, A.G, Irit, S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-02-19 | | Release date: | 2014-12-17 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
3BTP
 
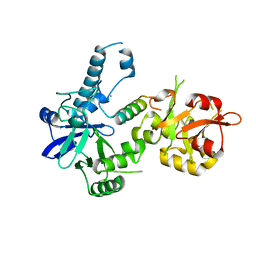 | | Crystal structure of Agrobacterium tumefaciens VirE2 in complex with its chaperone VirE1: a novel fold and implications for DNA binding | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, Protein virE1, ... | | Authors: | Dym, O, Albeck, S, Unger, T, Elbaum, M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2B5R
 
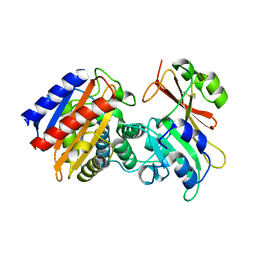 | | 1B Lactamase / B Lactamase Inhibitor | | Descriptor: | Beta-lactamase TEM, Beta-lactamase inhibitory protein | | Authors: | Rahat, O, Albeck, S, Meged, R, Dym, O, Screiber, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding Hot Spots in the TEM1-BLIP Interface in Light of its Modular Architecture.
J.Mol.Biol., 102, 2006
|
|
2F1O
 
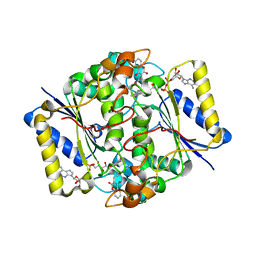 | | Crystal Structure of NQO1 with Dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Shaul, Y, Asher, G, Dym, O, Tsvetkov, P, Adler, J, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-11-15 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol.
Biochemistry, 45, 2006
|
|
3JYM
 
 | |
2B83
 
 | |
2RCI
 
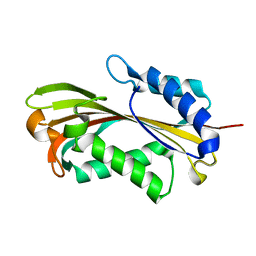 | |
2RKX
 
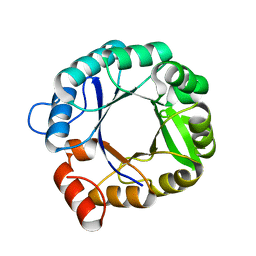 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Descriptor: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Authors: | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-10-18 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
3HQJ
 
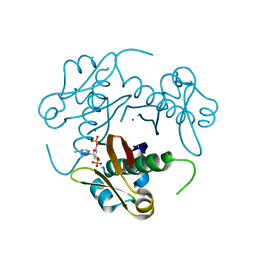 | | Structure-function analysis of Mycobacterium tuberculosis acyl carrier protein synthase (AcpS). | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase, MAGNESIUM ION | | Authors: | Dym, O, Albeck, S, Peleg, Y, Schwarz, A, Shakked, Z, Burstein, Y, Zimhony, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-06-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis.
J.Mol.Biol., 393, 2009
|
|
3JXV
 
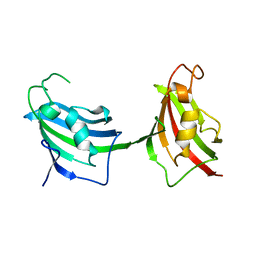 | |
3UZJ
 
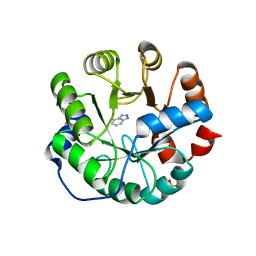 | | Designed protein KE59 R13 3/11H with benzotriazole | | Descriptor: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GKL
 
 | |
3BTN
 
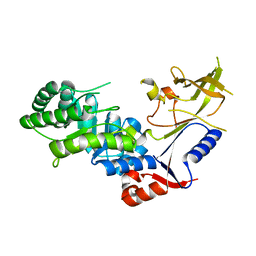 | | Crystal structure of antizyme inhibitor, an ornithine decarboxylase homologous protein | | Descriptor: | Antizyme inhibitor 1 | | Authors: | Dym, O, Unger, T, Albeck, S, Kahana, C, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.
Protein Sci., 17, 2008
|
|
3UTZ
 
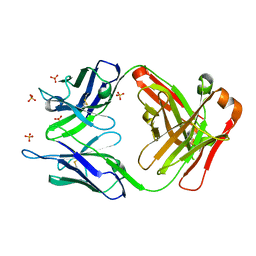 | | Endogenous-like inhibitory antibodies targeting activated metalloproteinase motifs show therapeutic potential | | Descriptor: | Metalloproteinase, heavy chain, light chain, ... | | Authors: | Sela-Passwell, N, Kikkeri, R, Dym, O, Rozenberg, H, Margalit, R, Arad-Yellin, R, Eisenstein, M, Brenner, O, Shoham, T, Danon, T, Shanzer, A, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-11-27 | | Release date: | 2011-12-14 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential.
NAT.MED. (N.Y.), 18, 2012
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | Descriptor: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
