3WR2
 
 | |
4XD7
 
 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
1SKY
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
1PFK
 
 | |
3WMR
 
 | | Crystal structure of VinJ | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, Proline iminopeptidase | | Authors: | Shinohara, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the amidohydrolase VinJ shows a unique hydrophobic tunnel for its interaction with polyketide substrates
Febs Lett., 588, 2014
|
|
2RPB
 
 | |
2EXD
 
 | | The solution structure of the C-terminal domain of a nfeD homolog from Pyrococcus horikoshii | | Descriptor: | nfeD short homolog | | Authors: | Kuwahara, Y, Ohno, A, Morii, T, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold.
Protein Sci., 17, 2008
|
|
5X18
 
 | | Crystal structure of Casein kinase I homolog 1 | | Descriptor: | Casein kinase I homolog 1, GLYCEROL, MALONIC ACID | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5X17
 
 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
8XGK
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Ihhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-(4-chlorophenyl)-2-fluoranyl-ethanoyl]amino]-3-[3-(2-cyano-2-methyl-propoxy)-4-methyl-phenyl]-2-methyl-propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
8XGV
 
 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (PPI) Inhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-[4-[(3-ethoxypyridin-2-yl)methyl]phenyl]-2-fluoranyl-ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
2LCF
 
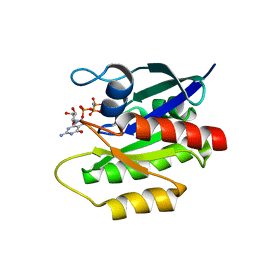 | | Solution structure of GppNHp-bound H-RasT35S mutant protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Araki, M, Shima, F, Yoshikawa, Y, Muraoka, S, Ijiri, Y, Nagahara, Y, Shirono, T, Kataoka, T, Tamura, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers.
J.Biol.Chem., 286, 2011
|
|
3AXB
 
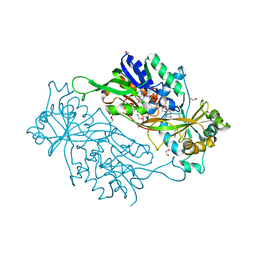 | | Structure of a dye-linked L-proline dehydrogenase from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, PROLINE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K, Hara, Y. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
4U4P
 
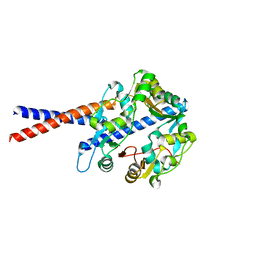 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1IW8
 
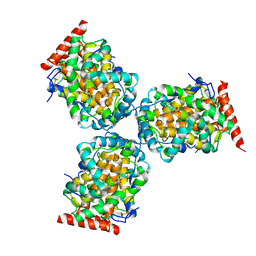 | | Crystal Structure of a mutant of acid phosphatase from Escherichia blattae (G74D/I153T) | | Descriptor: | SULFATE ION, acid phosphatase | | Authors: | Ishikawa, K, Mihara, Y, Shimba, N, Ohtsu, N, Kawasaki, H, Suzuki, E, Asano, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhancement of nucleoside phosphorylation activity in an acid phosphatase
PROTEIN ENG., 15, 2002
|
|
1J2F
 
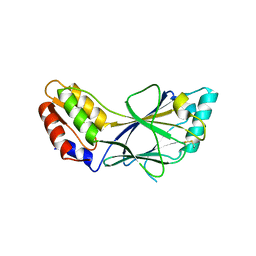 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
4U6K
 
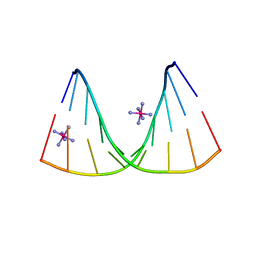 | | Crystal structure of DNA/RNA duplex containing 2'-4'-BNA-NC | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*(NCU)P*(NTT)P*CP*TP*TP*CP*TP*(NTT)P*(NCU))-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Kondo, J, Nomura, Y, Kitahara, Y, Obika, S, Torigoe, H. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a 2',4'-BNA(NC)[N-Me]-modified antisense gapmer in complex with the target RNA.
Chem.Commun.(Camb.), 52, 2016
|
|
4U6M
 
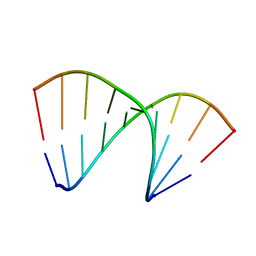 | | Crystal structure of DNA/RNA duplex obtained in the presence of Spermine | | Descriptor: | DNA (5'-D(*(5CM)P*TP*CP*TP*TP*CP*TP*TP*(5CM))-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Kondo, J, Nomura, Y, Kitahara, Y, Obika, S, Torigoe, H. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a 2',4'-BNA(NC)[N-Me]-modified antisense gapmer in complex with the target RNA.
Chem.Commun.(Camb.), 52, 2016
|
|
4U6L
 
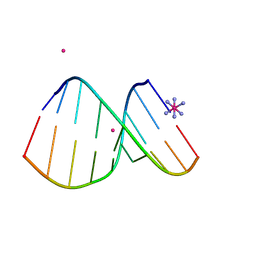 | | Crystal structure of DNA/RNA duplex obtained in the presence of [Co(NH3)6]Cl3 and SrCl2 | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*(5CM)P*TP*CP*TP*TP*CP*TP*TP*(5CM))-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3'), ... | | Authors: | Kondo, J, Nomura, Y, Kitahara, Y, Obika, S, Torigoe, H. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a 2',4'-BNA(NC)[N-Me]-modified antisense gapmer in complex with the target RNA.
Chem.Commun.(Camb.), 52, 2016
|
|
1V84
 
 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V83
 
 | | Crystal structure of human GlcAT-P in complex with Udp and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V82
 
 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
