8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
5XFY
 
 | | Crystal structure of a novel PET hydrolase S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XG0
 
 | | Crystal structure of a novel PET hydrolase from Ideonella sakaiensis 201-F6 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XFZ
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XH2
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with pNP from Ideonella sakaiensis 201-F6 | | Descriptor: | P-NITROPHENOL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
3KLX
 
 | |
4Y4M
 
 | | Thiazole synthase Thi4 from Methanocaldococcus jannaschii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Putative ribose 1,5-bisphosphate isomerase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R)-2,3,5-tris(oxidanyl)-4-oxidanylidene-pentyl] hydrogen phosphate | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
4Y4L
 
 | | Crystal structure of yeast Thi4-C205S | | Descriptor: | (2E)-2-[(2S,4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-yl]iminoethanoic acid, Thiamine thiazole synthase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
5XJC
 
 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5V56
 
 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
5V57
 
 | | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
1A83
 
 | | INTRAMOLECULAR I-MOTIF, NMR, 6 STRUCTURES | | Descriptor: | DNA (5'-(MCY)CTTTCCTTTACCTTTCC-3') | | Authors: | Han, X, Leroy, J.L, Gueron, M. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCT3CCT3ACCT3CC).
J.Mol.Biol., 278, 1998
|
|
7YKO
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
7YKQ
 
 | |
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
4QQR
 
 | |
3W28
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W29
 
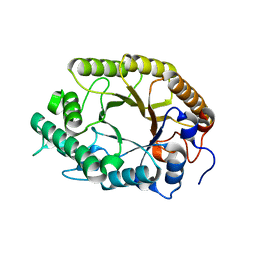 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W27
 
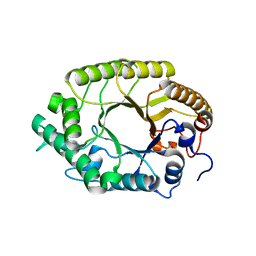 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W25
 
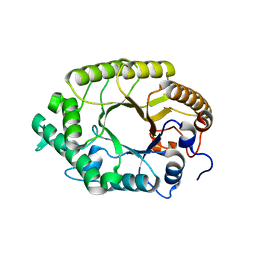 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W26
 
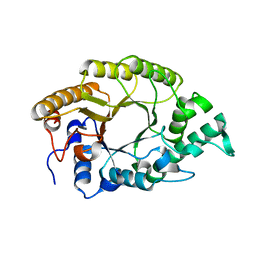 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
