3R21
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8RBM
 
 | |
8RB9
 
 | |
8RBQ
 
 | |
8RB8
 
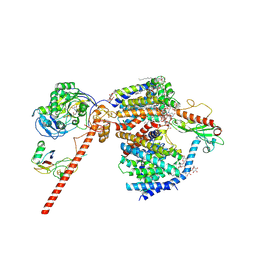 | | Cryo-EM structure of the NADH:ferredoxin oxidoreductase RNF from Azotobacter vinelandii, purified with 2-ME/TCEP, NADH added | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhang, L, Einsle, O. | | Deposit date: | 2023-12-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Architecture of the RNF1 complex that drives biological nitrogen fixation.
Nat.Chem.Biol., 20, 2024
|
|
7AQ8
 
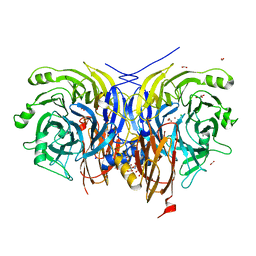 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y/D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ5
 
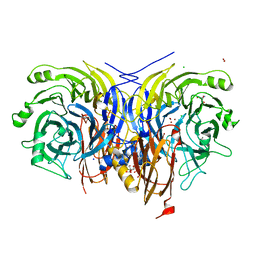 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583N | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ6
 
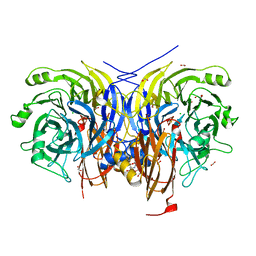 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583F | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ7
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ4
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583E | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ3
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583D | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7APY
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQA
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H382A | | Descriptor: | (dicuprio-$l^{3}-sulfanyl)-sulfanyl-copper, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
7AQ0
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A/S550A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ9
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583W | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ2
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
5XD6
 
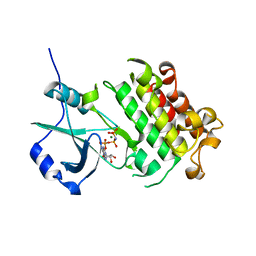 | | CARK1 phosphorylates ABA receptors | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase superfamily protein | | Authors: | Zhang, L, Lou, Z. | | Deposit date: | 2017-03-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | CARK1 phosphorylates ABA receptors
To Be Published
|
|
5HOQ
 
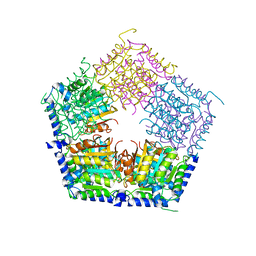 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
7YSE
 
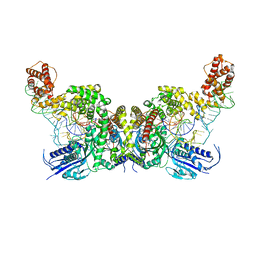 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
5BK7
 
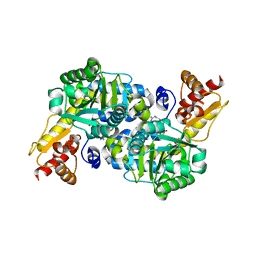 | | The structure of MppP E15A mutant soaked with the substrate L-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
5XPD
 
 | |
6IPB
 
 | |
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6C8T
 
 | | The structure of MppP soaked with the substrate L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
