4BJH
 
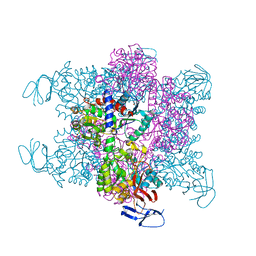 | | Crystal Structure of the Aquifex Reactor Complex Formed by Dihydroorotase (H180A, H232A) with Dihydroorotate and Aspartate Transcarbamoylase with N-(phosphonacetyl)-L-aspartate (PALA) | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, ASPARTATE CARBAMOYLTRANSFERASE, ... | | Authors: | Edwards, B.F.P, Martin, P.D, Grimley, E, Vaishnav, A, Fernando, R, Brunzelle, J.S, Cordes, M, Evans, H.G, Evans, D.R. | | Deposit date: | 2013-04-18 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mononuclear Metal Center of Type-I Dihydroorotase from Aquifex Aeolicus.
Bmc Biochem., 14, 2013
|
|
6PNZ
 
 | |
5DF5
 
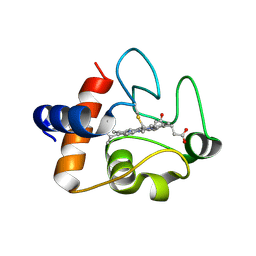 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
5C9M
 
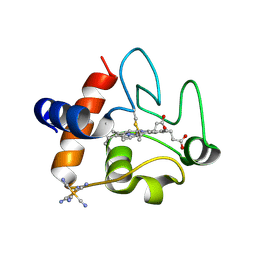 | | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution.
To Be Published
|
|
3D6N
 
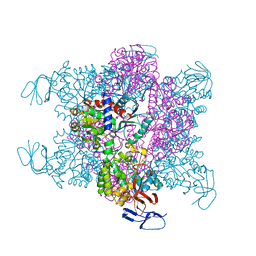 | | Crystal Structure of Aquifex Dihydroorotase Activated by Aspartate Transcarbamoylase | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, Dihydroorotase, ... | | Authors: | Edwards, B.F.P. | | Deposit date: | 2008-05-20 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.
Biochemistry, 48, 2009
|
|
8DVX
 
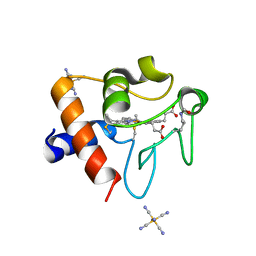 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
8DZL
 
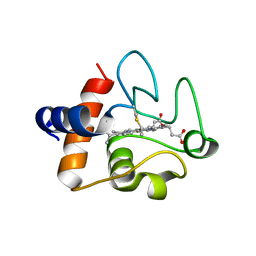 | | Structure of the K39Q mutant of rat somatic Cytochrome c at 1.36A | | Descriptor: | Cytochrome c, somatic, HEME C | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
5C0Z
 
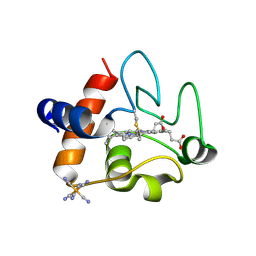 | | The structure of oxidized rat cytochrome c at 1.13 angstroms resolution | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1236 Å) | | Cite: | Serine-47 phosphorylation of cytochromecin the mammalian brain regulates cytochromecoxidase and caspase-3 activity.
Faseb J., 2019
|
|
1TVX
 
 | |
1YCP
 
 | |
1NAP
 
 | |
1MKX
 
 | |
1MKW
 
 | |
1SSC
 
 | | THE 1.6 ANGSTROMS STRUCTURE OF A SEMISYNTHETIC RIBONUCLEASE CRYSTALLIZED FROM AQUEOUS ETHANOL. COMPARISON WITH CRYSTALS FROM SALT SOLUTIONS AND WITH RNASE A FROM AQUEOUS ALCOHOL SOLUTIONS | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE A | | Authors: | De Mel, V.S.J, Doscher, M.S, Martin, P.D, Rodier, F, Edwards, B.F.P. | | Deposit date: | 1994-10-05 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1A0H
 
 | | THE X-RAY CRYSTAL STRUCTURE OF PPACK-MEIZOTHROMBIN DESF1: KRINGLE/THROMBIN AND CARBOHYDRATE/KRINGLE/THROMBIN INTERACTIONS AND LOCATION OF THE LINKER CHAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, MEIZOTHROMBIN | | Authors: | Martin, P.D, Malkowski, M.G, Box, J, Esmon, C.T, Edwards, B.F.P. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | New insights into the regulation of the blood clotting cascade derived from the X-ray crystal structure of bovine meizothrombin des F1 in complex with PPACK.
Structure, 5, 1997
|
|
1PLF
 
 | |
1SSB
 
 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Demel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
1SSA
 
 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
4CPV
 
 | |
1XRF
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | | Descriptor: | Dihydroorotase, SULFATE ION, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
1XRT
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex Aeolicus at 1.7 A Resolution | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
1VIT
 
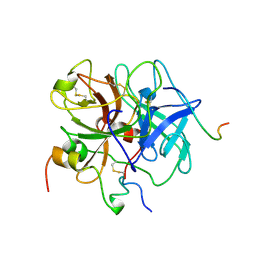 | | THROMBIN:HIRUDIN 51-65 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA THROMBIN, ... | | Authors: | Vitali, J, Edwards, B.F.P. | | Deposit date: | 1996-01-31 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a bovine thrombin-hirudin51-65 complex determined by a combination of molecular replacement and graphics. Incorporation of known structural information in molecular replacement.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1J9B
 
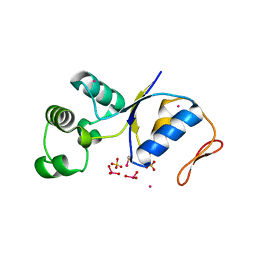 | | ARSENATE REDUCTASE+0.4M ARSENITE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-05-24 | | Release date: | 2001-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
1I9D
 
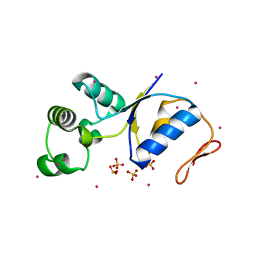 | | ARSENATE REDUCTASE FROM E. COLI | | Descriptor: | ARSENATE REDUCTASE, CESIUM ION, SULFATE ION, ... | | Authors: | Martin, P, Edwards, B.F. | | Deposit date: | 2001-03-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme.
Structure, 9, 2001
|
|
4SRN
 
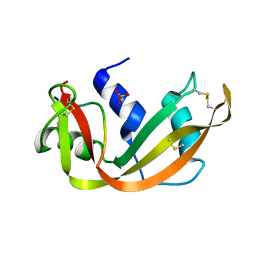 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
