1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULA
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1HIG
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN INTERFERON-GAMMA. | | Descriptor: | INTERFERON-GAMMA | | Authors: | Ealick, S.E, Cook, W.J, Vijay-Kumar, S, Carson, M, Nagabhushan, T.L, Trotta, P.P, Bugg, C.E. | | Deposit date: | 1991-10-03 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of recombinant human interferon-gamma.
Science, 252, 1991
|
|
1QCZ
 
 | |
5HFT
 
 | | Crystal structure of HpxW | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Ealick, S.E, Hicks, K.A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Biochemical and structural characterization of Klebsiella pneumoniae oxamate amidohydrolase in the uric acid degradation pathway.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2NSH
 
 | | E. coli PurE H45Q mutant complexed with nitro-AIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2NSL
 
 | | E. coli PurE H45N mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2NSJ
 
 | | E. coli PurE H45Q mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
2HS0
 
 | | T. maritima PurL complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HRY
 
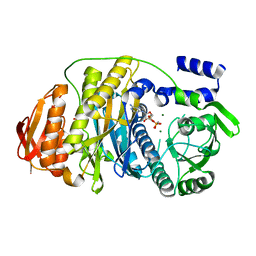 | | T. maritima PurL complexed with AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS4
 
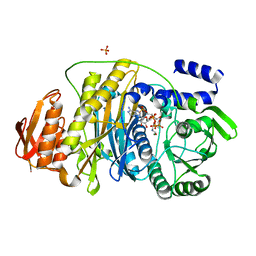 | | T. maritima PurL complexed with FGAR and AMPPCP | | Descriptor: | MAGNESIUM ION, N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS3
 
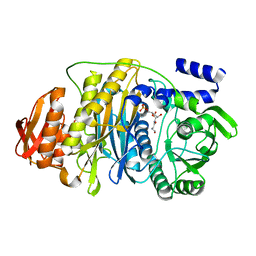 | | T. maritima PurL complexed with FGAR | | Descriptor: | N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HRU
 
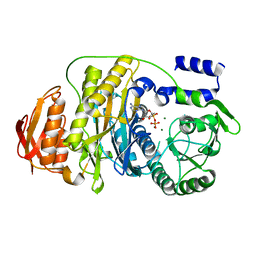 | | T. maritima PurL complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
1ZUD
 
 | | Structure of ThiS-ThiF protein complex | | Descriptor: | Adenylyltransferase thiF, CALCIUM ION, SODIUM ION, ... | | Authors: | Ealick, S.E, Lehmann, C. | | Deposit date: | 2005-05-30 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Escherichia coli ThiS-ThiF Complex, a Key Component of the Sulfur Transfer System in Thiamin Biosynthesis.
Biochemistry, 45, 2006
|
|
2QJI
 
 | | M. jannaschii ADH synthase complexed with dihydroxyacetone phosphate and glycerol | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2QQD
 
 | | N47A mutant of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AGMATINE, PYRUVIC ACID, ... | | Authors: | Ealick, S.E, Soriano, E.S. | | Deposit date: | 2007-07-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N47A and E109Q mutant proteins of pyruvoyl-dependent arginine decarboxylase from Methanococcus jannaschii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2QRO
 
 | | Human Deoxycytidine kinase dAMP, UDP, Mg ion product complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Deoxycytidine kinase, MAGNESIUM ION, ... | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structures of human deoxycytidine kinase product complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QQC
 
 | | E109Q mutant of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase subunit alpha, ... | | Authors: | Ealick, S.E, Soriano, E.S. | | Deposit date: | 2007-07-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N47A and E109Q mutant proteins of pyruvoyl-dependent arginine decarboxylase from Methanococcus jannaschii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2QRN
 
 | | Human Deoxycytidine kinase dCMP, UDP, Mg ion product complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Deoxycytidine kinase, MAGNESIUM ION, ... | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human deoxycytidine kinase product complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QJG
 
 | | M. jannaschii ADH synthase complexed with F1,6P | | Descriptor: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
2QRY
 
 | | Periplasmic thiamin binding protein | | Descriptor: | THIAMIN PHOSPHATE, Thiamine-binding periplasmic protein | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-30 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Similarities between Thiamin-Binding Protein and Thiaminase-I Suggest a Common Ancestor
Biochemistry, 47, 2008
|
|
2QJH
 
 | |
3D54
 
 | | Structure of PurLQS from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Formylglycinamide ribonucleotide amidotransferase, Phosphoribosylformylglycinamidine synthase 1, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2008-05-15 | | Release date: | 2008-07-22 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation.
Biochemistry, 47, 2008
|
|
1OV6
 
 | | M64V PNP + ALLO | | Descriptor: | 9-(6-DEOXY-BETA-D-ALLOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, P.W, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-25 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
1OTY
 
 | | Native PNP +ALLO | | Descriptor: | 6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, W.B, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-23 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
