8Q5P
 
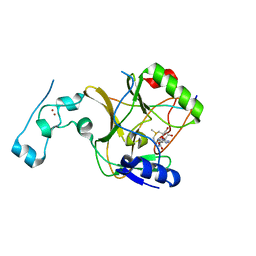 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
3D9W
 
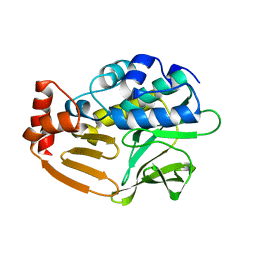 | | Crystal Structure Analysis of Nocardia farcinica Arylamine N-acetyltransferase | | Descriptor: | Putative acetyltransferase | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F, Martins, M, Dupret, J.M. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural characterization of the arylamine N-acetyltransferase from the opportunistic pathogen Nocardia farcinica
J.Mol.Biol., 383, 2008
|
|
3LNB
 
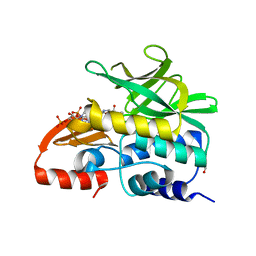 | | Crystal Structure Analysis of Arylamine N-acetyltransferase C from Bacillus anthracis | | Descriptor: | COENZYME A, FORMIC ACID, N-acetyltransferase family protein | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F. | | Deposit date: | 2010-02-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Bacillus anthracis arylamine N-acetyltransferase ((BACAN)NAT1) that inactivates sulfamethoxazole, reveals unusual structural features compared with the other NAT isoenzymes.
Febs Lett., 585, 2011
|
|
2BSZ
 
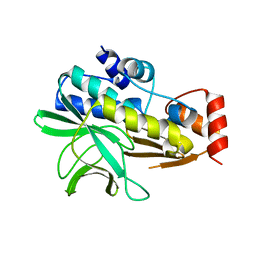 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4NV8
 
 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase F42W Mutant | | Descriptor: | Arylamine N-acetyltransferase | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NV7
 
 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase 1 In Complex With CoA | | Descriptor: | Arylamine N-acetyltransferase, COENZYME A | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IKP
 
 | | Crystal structure of human brain glycogen phosphorylase bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, brain form, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
5IKO
 
 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
1W4T
 
 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
6ZZ4
 
 | | Crystal structure of the PTPN2 C216G mutant | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Mechaly, A.E, Berthelet, J, Nian, Q, Parlato, M, Cerf-Bensussan, N, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a pathogenic mutant of human protein tyrosine phosphatase PTPN2 (Cys216Gly) that causes very early onset autoimmune enteropathy.
Protein Sci., 31, 2022
|
|
4DMO
 
 | | Crystal structure of the (BACCR)NAT3 arylamine N-acetyltransferase from Bacillus cereus reveals a unique Cys-His-Glu catalytic triad | | Descriptor: | 3-(1-methylpiperidinium-1-yl)propane-1-sulfonate, N-hydroxyarylamine O-acetyltransferase | | Authors: | Kubiak, X, Li de la Sierra-Gallay, I, Haouz, A, Weber, P, Rodrigues-Lima, F. | | Deposit date: | 2012-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and Biochemical Characterization of an Active Arylamine N-Acetyltransferase Possessing a Non-canonical Cys-His-Glu Catalytic Triad.
J.Biol.Chem., 288, 2013
|
|
4GUZ
 
 | | Structure of the arylamine N-acetyltransferase from Mycobacterium abscessus | | Descriptor: | Probable arylamine n-acetyl transferase | | Authors: | Kubiak, X, Li de la Sierra-Gallay, I, Haouz, A, Weber, P, Rodrigues-Lima, F. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of an arylamine N-acetyltransferase from the pathogen Mycobacterium abscessus: differences from other mycobacterial isoforms and implications for selective inhibition.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
