5HEV
 
 | |
4PDB
 
 | |
4WUH
 
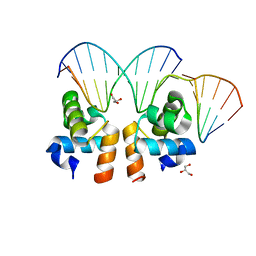 | | Crystal structure of E. faecalis DNA binding domain LiaR wild type complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
4WSZ
 
 | |
4WT0
 
 | |
4WUL
 
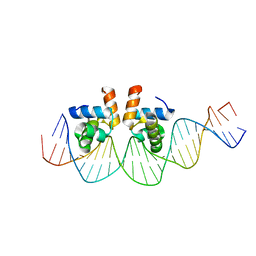 | |
4WU4
 
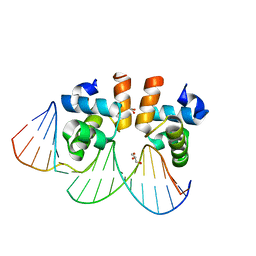 | | Crystal structure of E. faecalis DNA binding domain LiaRD191N complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*GP*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), GLYCEROL, ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
3S46
 
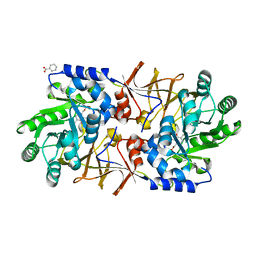 | | The crystal structure of alanine racemase from streptococcus pneumoniae | | Descriptor: | Alanine racemase, BENZOIC ACID | | Authors: | Im, H, Sharpe, M.L, Strych, U, Davlieva, M, Krause, K.L. | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of alanine racemase from Streptococcus pneumoniae, a target for structure-based drug design.
BMC MICROBIOL., 11, 2011
|
|
3P9U
 
 | | Crystal structure of TetX2 from Bacteroides thetaiotaomicron with substrate analogue | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein | | Authors: | Walkiewicz, K, Davlieva, M, Sun, C, Lau, K, Shamoo, Y. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of Bacteroides thetaiotaomicron TetX2: a tetracycline degrading monooxygenase at 2.8 A resolution.
Proteins, 79, 2011
|
|
3HA1
 
 | | Alanine racemase from Bacillus Anthracis (Ames) | | Descriptor: | ACETATE ION, Alanine racemase, CHLORIDE ION | | Authors: | Counago, R.M, Davlieva, M, Strych, U, Hill, R.E, Krause, K.L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of alanine racemase from Bacillus anthracis (Ames).
Bmc Struct.Biol., 9, 2009
|
|
6C0Z
 
 | |
6C84
 
 | |
6BWS
 
 | |
3FB4
 
 | | Crystal structure of adenylate kinase from Marinibacillus marinus | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Davlieva, M.G, Shamoo, Y. | | Deposit date: | 2008-11-18 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus marinus.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3H86
 
 | | Crystal structure of adenylate kinase from Methanococcus maripaludis | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Milya, D.G, Yousif, S. | | Deposit date: | 2009-04-28 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a trimeric archaeal adenylate kinase from the mesophile Methanococcus maripaludis with an unusually broad functional range and thermal stability.
Proteins, 78, 2009
|
|
