7PHS
 
 | |
5M38
 
 | |
1U0Q
 
 | |
1N8O
 
 | |
8BOZ
 
 | |
7NYJ
 
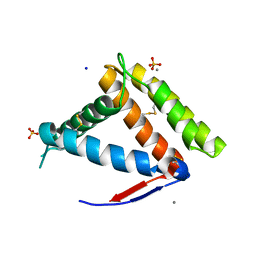 | | Structure of OBP1 from Varroa destructor, form P3<2>21 | | Descriptor: | CALCIUM ION, Odorant Binding Protein 1 from Varoa destructor, form P3<2>21, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7NZA
 
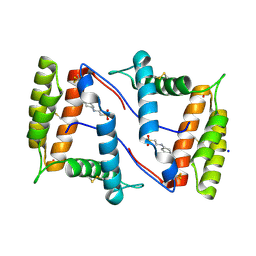 | | Structure of OBP1 from Varroa destructor, form P2<1> | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Odorant Binding Protein from Varroa destructor, form P2<1>, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
5NGH
 
 | |
2F0C
 
 | |
6EYY
 
 | |
6EYX
 
 | |
1FFD
 
 | |
1FFB
 
 | |
1FFC
 
 | |
1FFA
 
 | |
2A9M
 
 | |
2A9N
 
 | | A Mutation Designed to Alter Crystal Packing Permits Structural Analysis of a Tight-binding Fluorescein-scFv complex | | Descriptor: | 4-(2,7-DIFLUORO-6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)ISOPHTHALIC ACID, fluorescein-scfv | | Authors: | Cambillau, C, Spinelli, S, Honegger, A, Pluckthun, A. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation designed to alter crystal packing permits structural analysis of a tight-binding fluorescein-scFv complex.
Protein Sci., 14, 2005
|
|
1FFE
 
 | |
5M2J
 
 | | Complex between human TNF alpha and Llama VHH2 | | Descriptor: | Anti-(ED-B) scFV, Tumor necrosis factor | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5M2M
 
 | | Complex between human TNF alpha and Llama VHH3 | | Descriptor: | Llama nanobody VHH3, Tumor necrosis factor | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5M2Y
 
 | | Structure of TssK C-terminal domain from E. coli T6SS | | Descriptor: | TssK C | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A, Legrand, P. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
5M2I
 
 | | Structure of human Tumor Necrosis Factor (TNF) in complex with the Llama VHH1 | | Descriptor: | Tumor necrosis factor, VHH1 | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
5MWN
 
 | | Structure of the EAEC T6SS component TssK N-terminal domain in complex with llama nanobodies nbK18 and nbK27 | | Descriptor: | PHOSPHATE ION, Type VI secretion protein, llama nanobody raised against TssK, ... | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Legrand, P. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
6FVJ
 
 | | TesA a major thioesterase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Thioesterase, ... | | Authors: | Cambillau, C, Nguyen, V.S, Canaan, S. | | Deposit date: | 2018-03-03 | | Release date: | 2018-10-24 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Characterization of TesA, a Major Thioesterase Required for Outer-Envelope Lipid Biosynthesis in Mycobacterium tuberculosis.
J. Mol. Biol., 430, 2018
|
|
