1YT5
 
 | |
4D8M
 
 | | Crystal structure of Bacillus thuringiensis Cry5B nematocidal toxin | | Descriptor: | Pesticidal crystal protein cry5Ba | | Authors: | Fan, H, Hu, Y, Aroian, R.V, Ghosh, P, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-12-19 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Glycolipid Binding Properties of the Nematicidal Protein Cry5B.
Biochemistry, 51, 2012
|
|
3M93
 
 | |
3M94
 
 | | Complex crystal structure of Ascaris suum eIF4E-3 with m2,2,7G cap | | Descriptor: | ACETYL GROUP, Eukaryotic translation initiation factor 4E-binding protein 1, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Liu, W, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-03-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nematode eIF4E binding an m2,2,7G-Cap and its implications for translation initiation.
Nucleic Acids Res., 39, 2011
|
|
2BA2
 
 | | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae | | Descriptor: | Hypothetical UPF0134 protein MPN010 | | Authors: | Shin, D.H, Kim, J.-S, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-10-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae.
Protein Sci., 15, 2006
|
|
2MJP
 
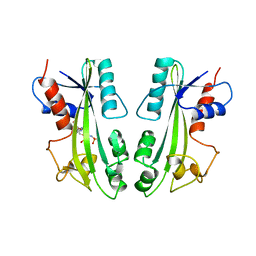 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
1DUS
 
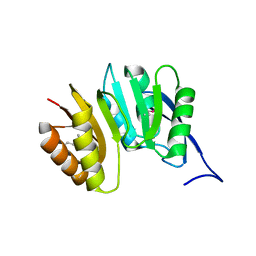 | | MJ0882-A hypothetical protein from M. jannaschii | | Descriptor: | MJ0882 | | Authors: | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
2AHD
 
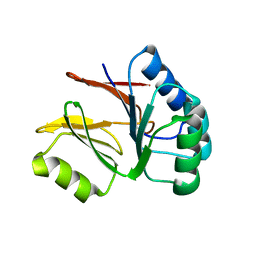 | |
2EIF
 
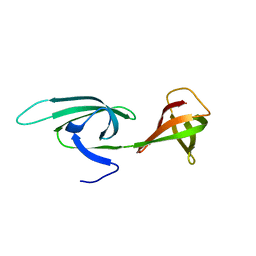 | | Eukaryotic translation initiation factor 5A from Methanococcus jannaschii | | Descriptor: | PROTEIN (EUKARYOTIC TRANSLATION INITIATION FACTOR 5A) | | Authors: | Kim, K.K, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1G8A
 
 | |
1FO5
 
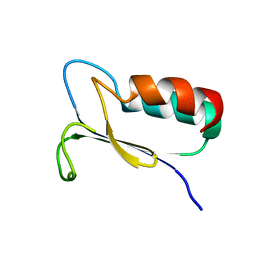 | | SOLUTION STRUCTURE OF REDUCED MJ0307 | | Descriptor: | THIOREDOXIN | | Authors: | Cave, J.W, Cho, H.S, Batchelder, A.M, Kim, R, Yokota, H, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-08-24 | | Release date: | 2001-04-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii.
Protein Sci., 10, 2001
|
|
1EKE
 
 | | CRYSTAL STRUCTURE OF CLASS II RIBONUCLEASE H (RNASE HII) WITH MES LIGAND | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RIBONUCLEASE HII | | Authors: | Lai, L.H, Yokota, H, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-03-07 | | Release date: | 2000-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of archaeal RNase HII: a homologue of human major RNase H
Structure, 8, 2000
|
|
1JX7
 
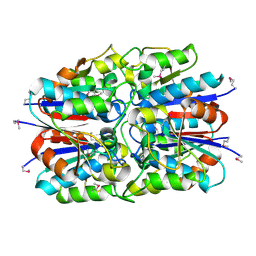 | | Crystal structure of ychN protein from E.coli | | Descriptor: | HYPOTHETICAL PROTEIN YCHN | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-09-05 | | Release date: | 2002-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a conserved hypothetical protein from Escherichia coli
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1F5S
 
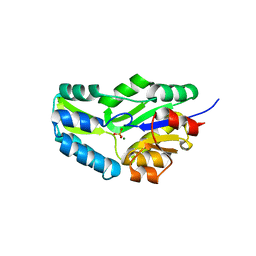 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
1FBN
 
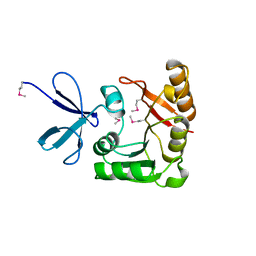 | | CRYSTAL STRUCTURE OF A FIBRILLARIN HOMOLOGUE FROM METHANOCOCCUS JANNASCHII, A HYPERTHERMOPHILE, AT 1.6 A | | Descriptor: | MJ FIBRILLARIN HOMOLOGUE | | Authors: | Wang, H, Boisvert, D, Kim, K.K, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-04-25 | | Release date: | 2000-04-26 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution.
EMBO J., 19, 2000
|
|
1G2I
 
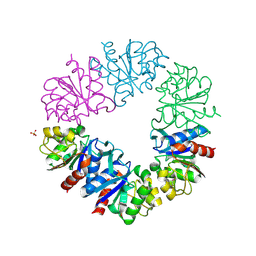 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1MJH
 
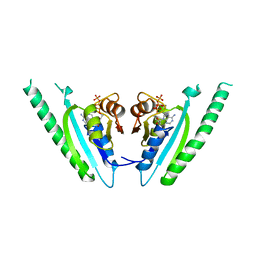 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | Authors: | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1LFP
 
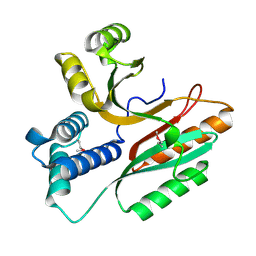 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LQL
 
 | | Crystal structure of OsmC like protein from Mycoplasma pneumoniae | | Descriptor: | osmotical inducible protein C like family | | Authors: | Choi, I.-G, Shin, D.H, Brandsen, J, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-05-10 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of a stress inducible protein from Mycoplasma pneumoniae at 2.85 A resolution
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1ILW
 
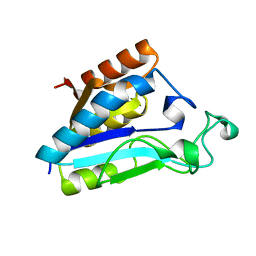 | |
1L7M
 
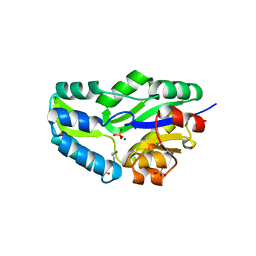 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7P
 
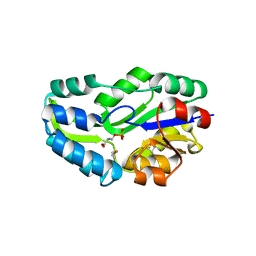 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
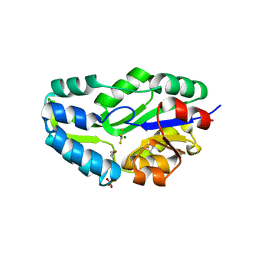 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L2F
 
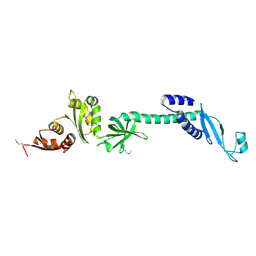 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1L7O
 
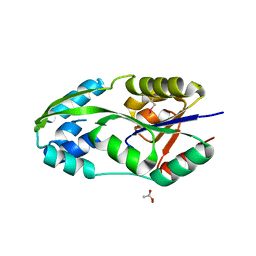 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
