6T9E
 
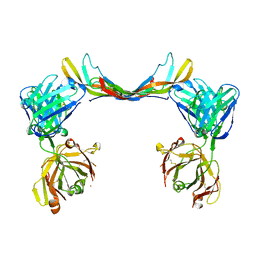 | | Crystal structure of a bispecific DutaFab in complex with human PDGF | | Descriptor: | DutaFab mat VH chain, DutaFab mat VL chain, Platelet-derived growth factor subunit B | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
6T9D
 
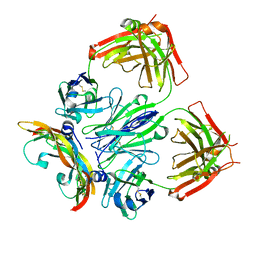 | | Crystal structure of a bispecific DutaFab in complex with human VEGF121 | | Descriptor: | VP mat DutaFab VH chain, VP mat DutaFab VL chain, Vascular endothelial growth factor A | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
4V7E
 
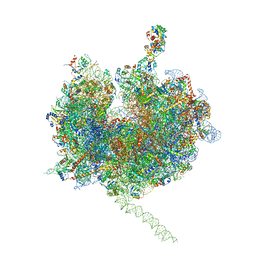 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | Authors: | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8RL2
 
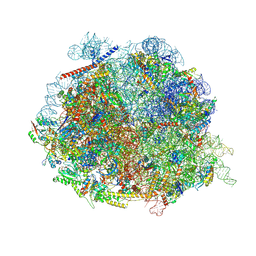 | | Human pre-60S - State 5 | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Denk, T, Beckmann, R. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Highly conserved ribosome biogenesis pathways between human and yeast revealed by the MDN1-NLE1 interaction and NLE1 containing pre-60S subunits.
Nucleic Acids Res., 53, 2025
|
|
2IX8
 
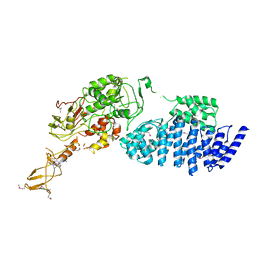 | | MODEL FOR EEF3 BOUND TO AN 80S RIBOSOME | | Descriptor: | ELONGATION FACTOR 3A | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
8BIP
 
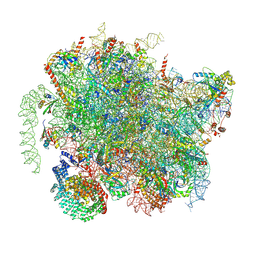 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BJQ
 
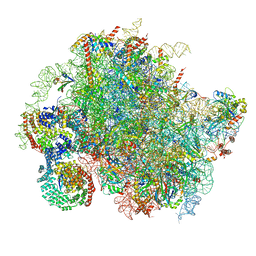 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6Q8Y
 
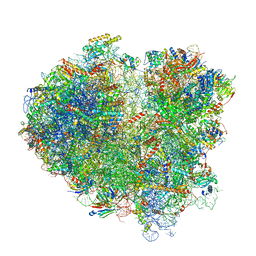 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6T83
 
 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7T
 
 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
5JCS
 
 | | CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Barrio-Garcia, C, Thoms, M, Flemming, D, Kater, L, Berninghausen, O, Bassler, J, Beckmann, R, Hurt, E. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling
Nat.Struct.Mol.Biol., 23, 2016
|
|
6TMF
 
 | | Structure of an archaeal ABCE1-bound ribosomal post-splitting complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kratzat, H, Becker, T, Tampe, R, Beckmann, R. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex.
Embo J., 39, 2020
|
|
5JTE
 
 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with A-, P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
5JU8
 
 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
5JPQ
 
 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
1YSH
 
 | | Localization and dynamic behavior of ribosomal protein L30e | | Descriptor: | 40S RIBOSOMAL PROTEIN S13, RNA (101-MER), RNA (28-MER), ... | | Authors: | Halic, M, Becker, T, Frank, J, Spahn, C.M, Beckmann, R. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Localization and dynamic behavior of ribosomal protein L30e
Nat.Struct.Mol.Biol., 12, 2005
|
|
9QIW
 
 | | Human pre-60S - State 3 | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Denk, T, Beckmann, R. | | Deposit date: | 2025-03-17 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Highly conserved ribosome biogenesis pathways between human and yeast revealed by the MDN1-NLE1 interaction and NLE1 containing pre-60S subunits.
Nucleic Acids Res., 53, 2025
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
4V6M
 
 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
