2AY0
 
 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
1TJ0
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) co-crystallized with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ1
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TIW
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | Descriptor: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ2
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | Descriptor: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
6ONU
 
 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
6VZ9
 
 | |
8DKG
 
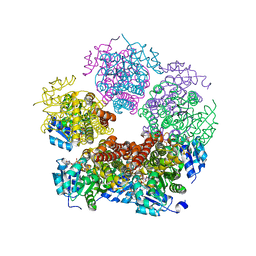 | | Structure of PYCR1 Thr171Met variant complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Isoform 3 of Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Meeks, K.R, Tanner, J.J. | | Deposit date: | 2022-07-05 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional Impact of a Cancer-Related Variant in Human Delta 1 -Pyrroline-5-Carboxylate Reductase 1.
Acs Omega, 8, 2023
|
|
3HAZ
 
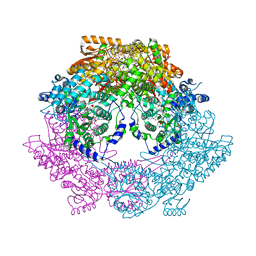 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ITG
 
 | |
2RBF
 
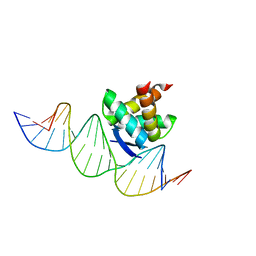 | | Structure of the ribbon-helix-helix domain of Escherichia coli PutA (PutA52) complexed with operator DNA (O2) | | Descriptor: | Bifunctional protein putA, DNA (5'-D(*DTP*DT*DTP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DCP*DAP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DGP*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DAP*DAP*DA)-3') | | Authors: | Tanner, J.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
J.Mol.Biol., 381, 2008
|
|
5KF7
 
 | |
5KF6
 
 | |
4JNZ
 
 | |
4JNY
 
 | |
4H6R
 
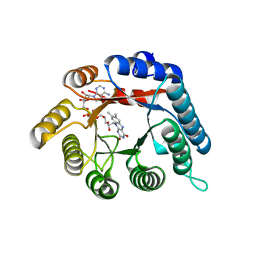 | | Structure of reduced Deinococcus radiodurans proline dehydrogenase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Proline dehydrogenase | | Authors: | Min, L, Tanner, J.J. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release.
Biochemistry, 51, 2012
|
|
4O8A
 
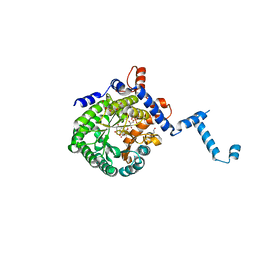 | |
4Q72
 
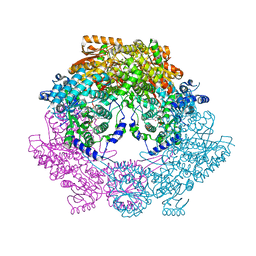 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D779Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Pemberton, T.A, Luo, M. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
4Q73
 
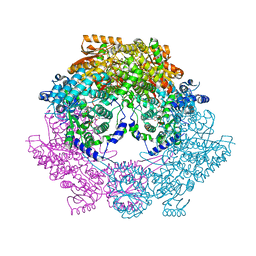 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D778Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
2JXH
 
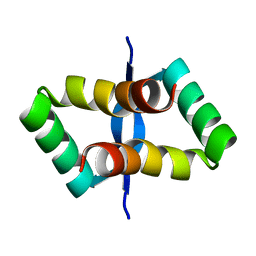 | |
2JXG
 
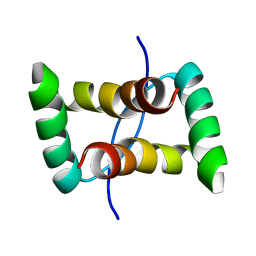 | |
2JXI
 
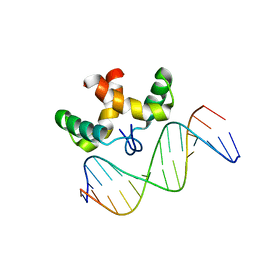 | | Solution structure of the DNA-binding domain of Pseudomonas putida Proline utilization A (putA) bound to GTTGCA DNA sequence | | Descriptor: | DNA (5'-D(*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DT)-3'), Proline dehydrogenase | | Authors: | Halouska, S, Zhou, Y, Becker, D, Powers, R. | | Deposit date: | 2007-11-19 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex
Proteins, 75, 2008
|
|
6D97
 
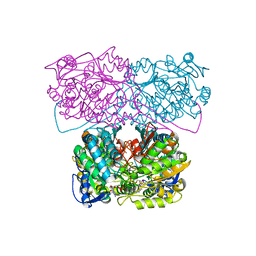 | |
6BSN
 
 | |
