1DL4
 
 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1S-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, DNA (5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Li, Z, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-01-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1S,2R,3S,4R)-N6-[1-(1,2,3,4-tetrahydro-2,3, 4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
1DJD
 
 | | THE SOLUTION STRUCTURE OF A NON-BAY REGION 11R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 8,9,10,11-TETRAHYDRO-BENZO[A]ANTHRACENE-8,9,10-TRIOL, DNA(5'-D(*CT*GT*GT*AT*CT*AT*AT*GT*AT*AT*G)-3'), DNA(5'-D(*CT*TT*TT*CT*TT*TT*GT*TT*CT*CT*G)-3') | | Authors: | Li, Z, Kim, H.Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-02 | | Release date: | 1999-12-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of a polycyclic aromatic hydrocarbon bay region ring in modulating DNA adduct structure: the non-bay region (8S,9R,10S, 11R)-N(6)-[11-(8,9,10,11-tetrahydro-8,9, 10-trihydroxybenz[a]anthracenyl)]-2' -deoxyadenosyl adduct in codon 61 of the human N-ras protooncogene
Biochemistry, 38, 1999
|
|
2QC3
 
 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
1I7V
 
 | | THE SOLUTION STRUCTURE OF A BAY REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*AP*(BZA)AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Li, Z, Tamura, P.J, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-03-10 | | Release date: | 2001-03-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1R,2S,3R,4S)-N6-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in the N-ras codon 61 sequence: DNA sequence effects
Biochemistry, 40, 2001
|
|
1YW5
 
 | | Peptidyl-prolyl isomerase ESS1 from Candida albicans | | Descriptor: | peptidyl prolyl cis/trans isomerase | | Authors: | Li, Z, Li, H, Devasahayam, G, Gemmill, T, Chaturvedi, V, Hanes, S.D, Van Roey, P. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Candida albicans Ess1 Prolyl Isomerase Reveals a Well-Ordered Linker that Restricts Domain Mobility
Biochemistry, 44, 2005
|
|
5KY4
 
 | | mouse POFUT1 in complex with mouse Notch1 EGF26 and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5L0T
 
 | | human POGLUT1 in complex with EGF(+) and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY9
 
 | |
5KY0
 
 | | mouse POFUT1 in complex with mouse Notch1 EGF12(D464G) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY2
 
 | |
5KXQ
 
 | | mouse POFUT1 in complex with GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5L0V
 
 | | human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-beta-D-glucopyranose, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KXH
 
 | | mouse POFUT1 in complex with mouse Factor VII EGF1 and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VII, GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5KY5
 
 | | mouse POFUT1 in complex with EGF(+) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EGF(+), GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5L0S
 
 | | human POGLUT1 in complex with Factor VII EGF1 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY8
 
 | |
5L0R
 
 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY3
 
 | |
5KY7
 
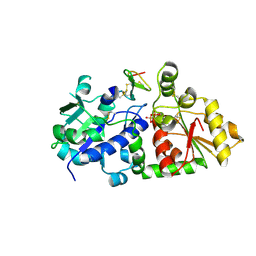 | | mouse POFUT1 in complex with O-glucosylated EGF(+) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EGF(+), GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
1QBY
 
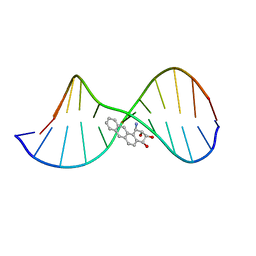 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP* G)-3' | | Authors: | Li, Z, Mao, H, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (-)-(1R,2S,3R, 4S)-N6-[1-benz[a]anthracenyl]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
4ETX
 
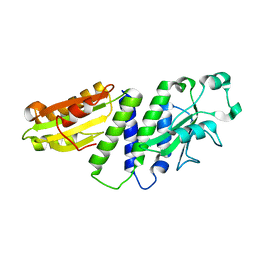 | |
4EUV
 
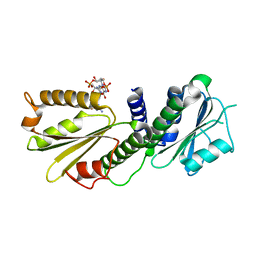 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1, in complex with c-di-GMP, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-25 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
3KYS
 
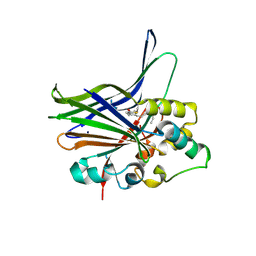 | | Crystal structure of human YAP and TEAD complex | | Descriptor: | 65 kDa Yes-associated protein, Transcriptional enhancer factor TEF-1 | | Authors: | Li, Z, Zhao, B, Wang, P, Chen, F, Dong, Z, Yang, H, Guan, K.L, Xu, Y. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-23 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the YAP and TEAD complex
Genes Dev., 24, 2010
|
|
8W9R
 
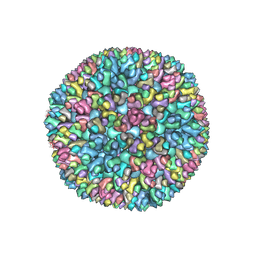 | | Structure of Banna virus core | | Descriptor: | VP10, VP8, Vp2 | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
