1BOY
 
 | |
4UD5
 
 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Plasticity of Cid1 Provides a Basis for its Distributive RNA Terminal Uridylyl Transferase Activity.
Nucleic Acids Res., 43, 2015
|
|
4V2B
 
 | | rat Unc5D Ig domain 1 | | Descriptor: | PROTEIN UNC5D | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4CSA
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4LP7
 
 | |
4V2A
 
 | | human Unc5A ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4V2D
 
 | | FLRT2 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
5FXU
 
 | | Crystal Structure of Puumala virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE POLYPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
5FN7
 
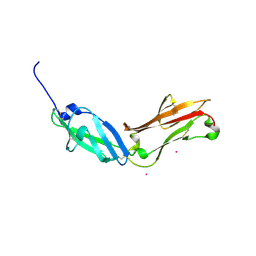 | | Crystal structure of human CD45 extracellular region, domains d1-d2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
1HNF
 
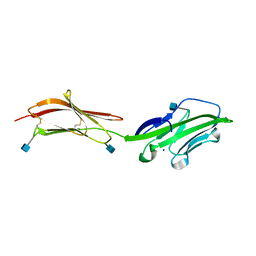 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
1QFT
 
 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
2X11
 
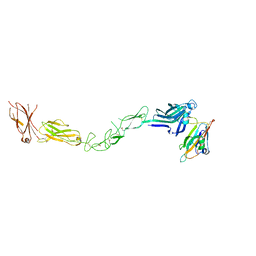 | | Crystal structure of the complete EphA2 ectodomain in complex with ephrin A5 receptor binding domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR 2, EPHRIN-A5 | | Authors: | Seiradake, E, Harlos, K, Sutton, G, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.83 Å) | | Cite: | An Extracellular Steric Seeding Mechanism for Eph-Ephrin Signalling Platform Assembly
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XBO
 
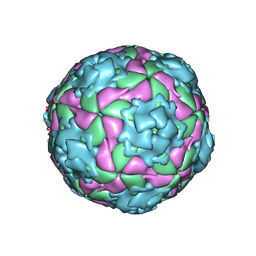 | | Equine Rhinitis A Virus in Complex with its Sialic Acid Receptor | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, P1 | | Authors: | Fry, E.E, Tuthill, T.J, Harlos, K, Walter, T.S, Rowlands, D.J, Stuart, D.I. | | Deposit date: | 2010-04-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of Equine Rhinitis a Virus in Complex with its Sialic Acid Receptor.
J.Gen.Virol., 91, 2010
|
|
5MQW
 
 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
5MGT
 
 | | Complex of human NKR-P1 and LLT1 in deglycosylated forms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, CHLORIDE ION, ... | | Authors: | Blaha, J, Skalova, T, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
5MGR
 
 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
5E8C
 
 | | pseudorabies virus nuclear egress complex, pUL31, pUL34 | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Zeev-Ben-Mordehai, T, Cheleski, J, Whittle, C, El Omari, K, Harlos, K, Hagen, C, Klupp, B, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Cell Rep, 13, 2015
|
|
4UOI
 
 | | Unexpected structure for the N-terminal domain of Hepatitis C virus envelope glycoprotein E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GENOME POLYPROTEIN | | Authors: | El Omari, K, Iourin, O, Kadlec, J, Harlos, K, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-08-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unexpected Structure for the N-Terminal Domain of Hepatitis C Virus Envelope Glycoprotein E1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8CKM
 
 | | Semaphorin-5A TSR 3-4 domains | | Descriptor: | Semaphorin-5A | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKG
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sulfate | | Descriptor: | SULFATE ION, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKK
 
 | | Semaphorin-5A TSR 3-4 domains in complex with nitrate | | Descriptor: | NITRATE ION, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKL
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-2)-1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
6THG
 
 | | Cedar Virus attachment glycoprotein (G) in complex with human ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.074 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
6THB
 
 | | Receptor binding domain of the Cedar Virus attachment glycoprotein (G) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
1GP3
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
